Effect of MSTN Propeptide and shRNA Co-expression Vector on Proliferation of Skeletal Muscle Satellite Cells
2014-03-07FengLinheWangXinLuMingTongHuiliLiShufengandYanYunqin
Feng Lin-he, Wang Xin, Lu Ming, Tong Hui-li, Li Shu-feng, and Yan Yun-qin
College of Life Sciences, Northeast Agricultural University, Harbin 150030, China
Effect of MSTN Propeptide and shRNA Co-expression Vector on Proliferation of Skeletal Muscle Satellite Cells
Feng Lin-he, Wang Xin, Lu Ming, Tong Hui-li, Li Shu-feng, and Yan Yun-qin*
College of Life Sciences, Northeast Agricultural University, Harbin 150030, China
Myostatin (MSTN) is a negative regulator of skeletal muscle growth, in order to study the effect of inhibition MSTN expression on the proliferation of bovine skeletal muscle satellite cells, we constructed co-expression vector pcDNA3.1-Pro-MSTNshRNA, transfected it into muscle satellite cells by Liposome 2000, and detected cell proliferation changes by CCK-8 method and flow cytometry after 48 h. The expressions of P21 and CDK2 were detected by Western blot and real-time PCR. The results showed that the cell vitality of experimental groups significantly increased than that of the negative control, and cells in S phase also increased significantly (P<0.05). After knocked down MSTN gene, P21 expression decreased (P<0.05), but CDK2 gene expression increased (P<0.05). These results indicated that MSTN gene expression was associated with P21 and CDK2, the proliferation of skeletal muscle satellite cells could be promoted while MSTN was inhibited, which provided a theoretical basis for the study on transgenic cattle.
myostatin, cell proliferation, flow cytometry, expression vector
Introduction
Myostatin (MSTN), also known as growth differentiation factor (GDF-8), which belongs to the transforming growth factor super family (TGF), is a negative regulator of muscle growth. Variation in MSTN gene has been associated with variation in muscularity in many animals including bovine, which exists in various tissues of animals. MSTN precursor protein is composed of three components: signal peptide, propeptide and C-terminal mature peptide (McPherron et al., 1997). The precursor protein is proteolytically cleaved at the RSRR (263) site to produce mature peptide. Two mature peptides with two disulfide binding, propeptide and mature peptide binding exists in the blood with covalent bond, besides the N-terminal propeptide can specifically inhibit the activity of mature peptide (Wolfman et al., 2003), and myostatin mature peptide can inhibit the proliferation of skeletal muscle cells, and then inhibit the growth of muscle. The cell proliferation relational protein synthesis is restrained by myostatin in C2C12 cells (McPherron et al., 1997; Gill et al., 2009; McCroskery et al., 2003). Myostatin null mouse exhibits two-fold increase in muscle mass compared with wild-type mice. Similarly experiments showed that content of muscle in transgenic mice, which was compared with MSTN propeptide, could over-express and increase by 22%-44% and the growth rate increases by 17%-30%, compared with control group mouse (Yang et al., 2001; Whittemore et al., 2003).
RNAi has been becoming an indispensable tool in functional genomics research, an effective methodof sequence-specific gene silencing, and specifically silenced targeted gene at mRNA level (Magee et al., 2006). It's a long term method to inhibit specific gene expression. Four MSTN shRNA sequences designed by ambion sofware were connected into the downstream of Pol III or Pol II promoter and constructed into pGenesil interference vector (Ji et al., 1998). Three shRNA vectors were designed based on the bovine MSTN gene conserved region, and we screened the most effective vector, then constructed co-expression vector of MSTN propeptide and MSTN shRNA. Subsequently, the vector was transfected into bovine skeletal muscle satellite cells. We explored that the coexpression vector promoted proliferation of bovine skeletal muscle satellite cell and had effects on generelated proliferation. This study focused on the effects on cell proliferation produced by the co-expression vector, and also provided a theoretical basis for the subsequent transgenic bovine experiment.
Materials and Methods
Materials
PMD18T vector was purchased from Dalian TaKaRa Biotechnology Co., Ltd.; pGenesil1.0, pcDNA3.1 (+) and pEGFP-C1were saved by our laboratory.
Tools enzymes, kits and other reagents
LA Taq DNA polymerase, PrimeScript™ RT reagent kit with gDNA Eraser (Perfect Real Time), restriction enzyme were purchased from Dalian TaKaRa Biotechnology Co., Ltd.; DMEM and serum were purchased from Gibco Co., Ltd.; GAPDH mouse monoclonal antibody was from Sata Co., Ltd.; P21, CDK2 and HRP labeled goat anti rabbit were from Bio Syntesis Co., Ltd.; cell cycle and apoptosis analysis kit were purchased from Beyotime Co., Ltd.; Flow Cytometer.
Methods
Design and identification of MSTN shRNA
According to Tush1 principle of siRNA of interference se-quence, three MSTN-specific interference sequences were designed and synthesized by Shanghai Biotechnology Company. After annealing, they were cloned into the pGenesil1.0 vector and transformed into E. coli DH5α. Picked a positive monoclonal colony, and identified by restriction enzyme and sequencing (Invitrogen Biotechnology).
Construction of MSTN gene eukaryotic expression vector
According to MSTN mRNA sequences of bovine MSTN gene published by GenBank, cDNA of skeletal muscle cell was used as template, designed specific primers to obtain MSTN sequence. The amplified products by PCR were connected with clone vector and sequenced by Invitrogen Biotechnology. The correct sequences were digested by the specific restriction endonuclease, and these products digested by the restriction enzyme were cloned into expression vectors and transformed into E.coli DH5α. Then picked a positive monoclonal clone, the plasmids were extracted and identified. The correct plasmid digested by restriction enzyme plasmid was named as pEGFPMSTN.
Construction of bovine eukaryotic co-expression vector
The co-expression vector pcDNA3.1-Pro-MSTNsh-RNA was constructed according to the methods described previously. Polyclonal specific enzyme linker sequence was designed and synthesized, then cloned into pEGFP-C1 and named as pEGFP-JT. pEGFP-JT and pGenesil-MSTNshRNA were digested by Sal I and Hind III restriction enzymes. The targeted DNA fragment retrieved by agarose gel was cloned into expression vector; then extracted the plasmids, and designated as pEGFP-MSTNshRNA. pEGFP-MSTN-shRNA vector and pcDNA3.1-Pro vectors were digested by Mlu I and Mfe I restriction enzymes, the fragments retrieved by agarose gel were cloned into vector fragments, and transformed into E. coli DH5α. Picked the positive monoclonal colonies, and the correct plasmid was named as pcDNA3.1-Pro-MSTNshRNA.
lnterference efficiency of bovine MSTNshRNA
The plasmid was prepared by Edo-Free Plasmid Kit. Plasmid transfection reagent was Lipofectamine 2000 Reagent. pGenesil-MSTNshRNA vector was cotransfected with pEGFP-MSTN into skeletal muscle satellite cells clutured in 6-cell plates by Lipofectamine 2000. Cell was cultured under a humidified atmosphere of 5% CO2at 37℃. Total RNA from primary cultured cells was extracted using Trizol (Invitrogen, Carlsbad, CA, USA), then further purified by phenol/chloroform extraction. Expression of MSTN, β-actin, P21and CDK2 were detected by SYBR Green Quantitative RT-PCR. Real-time RTPCR was performed in an ABI 7300 system (Applied Biosystems, UK). Each real-time RT-PCR was amplified in triplicate, the result was expressed as X±S, and the related ratios were calculated according to CT 2-ΔΔCtmethod. All data were analyzed by using statistical software SPSS17.0, P<0.05 or P<0.01 represented significant difference or extremely significance, which had a statistical meaning. Primer sets are displayed in Table 1.

Table 1 Primer sequence and fragment size
Western blot
After transfected for 48 h, cells total protein was extracted from each plate, and separated by SDS-PAGE on 10% gels. Used 1 : 1 000 rabbit monclonal antibody as primary antibody, and then washed membranes by PBS three times, which were incubated with a 1 : 2 000 dilution of HRP labeled goat anti rabbit as second antibody for 1 h. Finaly, used DAB to display colors.
Cell viability detected by CCK8 reagent
After transfected for 48 h, cells were prepared single cell suspension with the culture medium including 20% fetal bovine serum and 10% horse serum, and inoculated in 96 cell plates in 1.0×105cells • mL-1, added CCK8 reagent after 48 h, each plate OD value was detected in 450 nm wavelength by enzyme-labeled meter, and each group of five parallels was repeated three times.
Detection of cell cycle by flow cytometry
After transfected for 48 h, the cells were blowed into single cell suspension by growth medium, the cells were fixed with 70% ethanol for 10 h, discarded 70% ethanol, and washed two times with PBS, added 500 uL PI reagent and 10 uL RNAase to the cell precipitation, then blowed into single cell suspension, and detected cell proportions in S phase by flow cytometry.
Results
Design of bovine MSTNshRNA target sequences
Bovine myostatin small hairpin sequence was designed by Invitrogen ambion siRNA design software. Three potentially effective interference fragments were selected. The targeted sequences are shown in Table 2.
Construction of MSTN eukaryotic expression vector
Using bovine skeletal muscle satellite cell cDNA as the template, the targated products, whose size was 1 265 bp, were amplified by specific primers. The products were consistent with the expected size, but only one base was difference from the targeted sequence from GenBank. A bovine myostatin-expression plasmid pEGFP-MSTN was successfully constructed. MSTN expression vector construction process is shown in Fig. 1, and co-expression vector construction process is shown in Fig. 2.

Table 2 shRNA target sequence

Fig. 1 Construction of MSTN expression vector
Inhibition of MSTN gene mRNA expression using shRNA expression vectors
pEGFP-MSTN was co-transfected with MSTNshRNA into bovine fetal fibroblast cells. The best interference efficiency vector was selected by real-time PCR according to CT value, the relative expression of MSTN gene in each group was calculated, and the results are shown in Fig. 3. The cells transfected after 48 h achieved relative interference efficiency, pGenesilMSTNsh1 interference efficiency was 35.3%, pGenesilMSTNsh2 interference efficiency was only 15.0%, and pGenesil MSTNsh3 interference efficiency was 65.3%. The results showed that pGenesilMSTNsh3 interference efficiency was obviously remarkable, and then they were used to build the co-expression vector.
Detection of P21 and CDK2 expressions by real-time PCR
The total RNA was respectively prepared from each experimental group cells which were co-transfected with pGenesil-control, pcDNA3.1-Pro, pGenesil-MSTN-shRNA, and pcDNA3.1-Pro-MSTNshRNA. The relative P21 and CDK2 gene expressions are shown in Fig. 4.
The experimental result showed that P21 gene expression of pcDNA3.1-Pro-MSTNshRNA, pcDNA3.1-Pro and pGenesil-MSTNshRNA groups was lower than that of control (P<0.05), and P21 expression of pcDNA3.1-Pro-MSTNshRNA group was lower than that of other two experimental groups (P<0.05). With the reduction of MSTN gene expression, CDK2 gene mRNA expression level of pcDNA3.1-Pro-MSTN-shRNA, pcDNA3.1-Pro and pGenesil-MSTNshRNA groups was higher than that of the negative control (P<0.05), and CDK2 gene mRNA expression level of pcDNA3.1-Pro-MSTNshRNA was higher than that of other two experimental groups.
Protein expression levels of experimental group detected by Western blot
Total protein was prepared from different groups ofcells transformed with various eukaryotic expression vectors. The results are shown in Fig. 5. P21 protein expressions of pcDNA3.1-Pro-MSTN-shRNA were lower than those of the control group and other two experimental groups. CDK2 protein expression level of the cell transfected pcDNA3.1-Pro-MSTNshRNA group was higher than that of the negative control group and other two experimental groups.
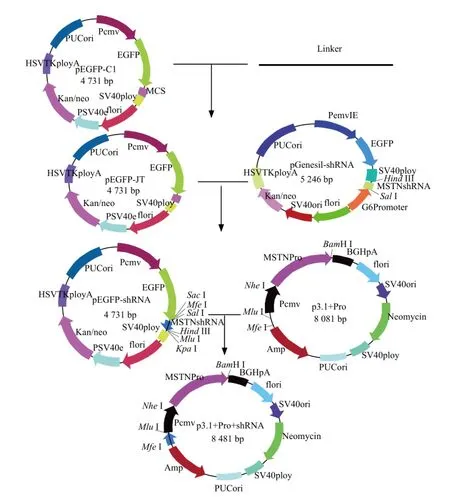
Fig. 2 Construction of co-expression vector
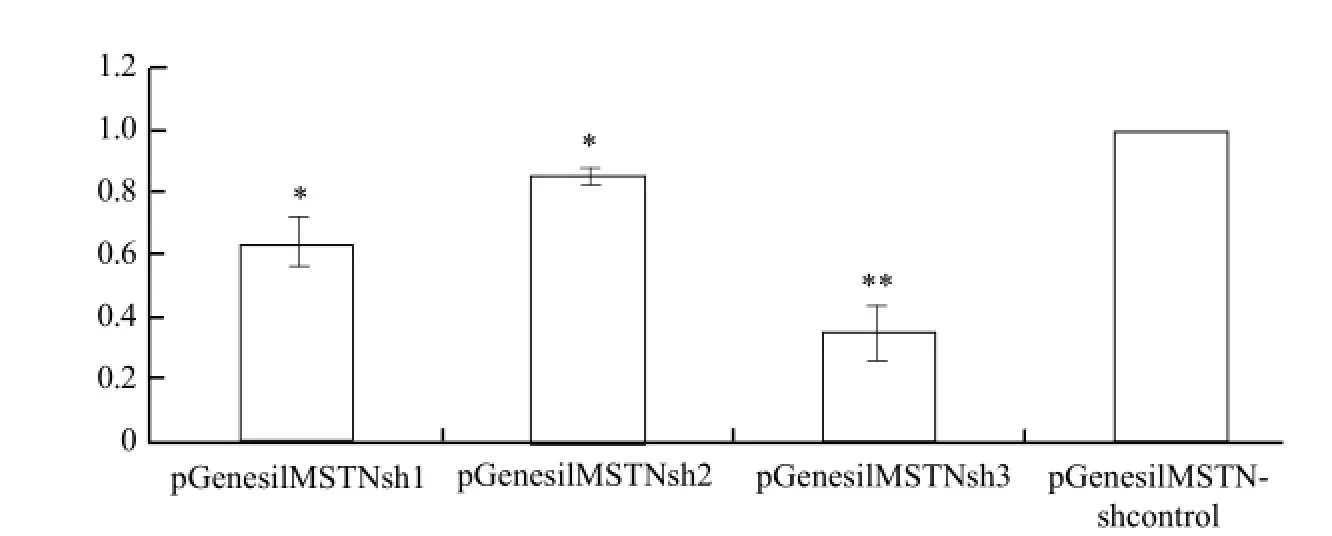
Fig. 3 Selection for effective shRNA by real-time PCR

Fig. 4 P21 and CDK2 expression analyzed by real-time PCR
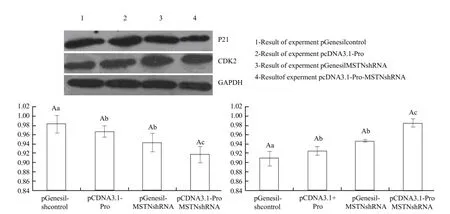
Fig. 5 Detection of cell P21 and CDK2 mRNA expression levels by real-time PCR
Cells proliferating ability detected by enzyme mark instrument
Bovine skeletal muscle satellite cells were transfected with experimental vectors. The cell viability after 48 h was detected by CCK-8 reagent kit. The cells' viability was calculated through OD value (Fig. 6).
It showed that the absorbance value of three experimental groups was significantly higher than that of the negative control (P<0.05), and the absorbance value of pcDNA3.1-Pro-MSTNshRNA group was higher than that of pcDNA3.1-Pro and pGenesil-MSTNshRNA groups (P<0.05). These results indicated that the co-expression vector had a cumulative effect on the cells' proliferating ability.

Fig. 6 Cell viability detected by enzyme mark instrument
Detection of cell cycle changes after transfected plasmids by flow cytometry
Fig. 7 showed that cell proportions in S phase of experimental groups was higher than that of the negativecontrol (P<0.05), and cell proportions in S phase with pcDNA3.1-Pro-MSTNshRNA group was higher than that of pcDNA3.1-Pro and pGenesil-MSTNshRNA groups (P<0.05). It indicated that the co-expression vector had a significant effect on promoting proliferation ability of bovine skeletal muscle satellite cells.

Fig. 7 Cell proportions in S phase of cell cycle
Discussion
Currently, shRNA expression vector promoters are mainly H1 and U6 of type Pol III, which have the characteristics of simple structures, fixed transcription sites, less burdensome sequences, and terminating transcription with continuous 5-6 T. In addition, type Pol II promoters can efficiently start small RNA transcription, such as CMV promoter, SPC promoter, and CMV promoter can particularly drive small RNA transcription of effective target sequence containing a plurality of T. Studies showed that many tissue-specific Pol II promoters can drive shRNA transcription with different intensities (Xia et al., 2006). Dai et al. (2011) used CMV promoter and human keratin 14 gene promoter of hair follicle-specific expression to achieve eGFP-shRNA fusion transcript. Multiple experiment groups studied shRNA transcription drived by tissuespecific promoters, which provided experimental evidence for gene knockdown specific organization to achieve tissue-specific target gene silencing. Studies showed that aiming at a signaling pathway and multiple cytokines, we could design multiple targeting shRNA, and simultaneously design two complete coding regions to construct into the same vector to achieve the over-expression of two target genes, and four shRNA efficient and silenced target gene vectors, which provided a new method for study on signaling pathways.
MSTN is a member of transforming growth factor superfamily, and is a secreted glycoprotein expressed widely in skeletal muscles, which can affect the animal muscle contents through regulating the target gene expression. Experimental results showed that inhibiting MSTN gene expression could promote muscle growth and development (Zhu et al., 2000). At present, the common method is to add myostatin antagonists, over-expression of myostatin propeptide, and oligo RNA nucleotide sequence. Using siRNA or shRNA technology to inhibit MSTN gene expression has been widely reported, shRNA has relatively long timespecific silencing gene expression, and then quickly gained RNAi adult animals, which provided a new method for the study of gene function.
MSTN is widely expressed in skeletal muscle cells, and less expressed in fat tissue. A lot of researches showed that MSTN specifically regulated skeletal muscle satellite cell proliferation, when muscle damages occurred, in the role of the relevant activated factors, skeletal satellite cells would rapidly proliferate, which is closely related to MSTN (Taylor, 2001; Thomas et al., 2000). In addition, MSTN gene expression has a negative regulatory role for muscle development during the embryonic development, but excessive inhibiting MSTN activity will generate negative impacts. MSTN null mutant animals exhbited reduced reproductive capacity and excessive heat during exercise (Gill et al., 2009; Chupin, 1982). These phenomena might associate with the high numbers of muscle fibers. In a certain extent, moderate inhibiting gene expression is particularly important, while siRNA inhibits target gene expression in a very broad range, so this advantage is of great significance to study transgenic animals (Patel and Amthor, 2005).
Conclusions
The experiment successfully constructed co-expression vector pcNDA3.1-Pro-MSTNshRNA eukaryotic expression vector, and detected the effects on bovine skeletal muscle satellite cell proliferation. It showed that the proliferation of bovine skeletal muscle satellite cells transfected with the co-expression vector was significantly higher than those of other two experimental groups and the control group, and it indicated that the co-expression vector played the dual role that enhanced the ability of cell proliferation. This study laid the theoretical foundation for the subsequent transgenic cattle research.
Chupin D. Analysis of reproduction problems in double muscle females. In: King J W B, Menissier F. Muscle hypertrophy of genetic origin and its use to improve beef production. Dordrecht: Kluwer, 1982.
Dai R, Shen S J, Wan P C, et al. 2011. shRNAs driven by K14 promoter induce tissue-specific RNA interference. Hereditas, 33(7): 757-762.
Gill J L, Bishop S C, McCorquodale C, et al. 2009. Associations between the 11-bp deletion in the myostatin gene and carcass quality in Angus-sired cattle. Anim Genet, 40(1): 97-100.
Ji S, Losinski R L, Cornelius S G, et al. 1998. Myostatin expression in porcine tissues: tissue specificity and developmental and postnatal regulation. Am J Physiol Regul Integr Comp Physiol, 275(4pt2): R1265-R1273.
John S, John G, Daniel B. 1997. TGF-β latency: biological significance and mechanisms of activation. Stem Cells, 15: 190-197.
Magee T R, Artaza J N, Ferrini M G, et al. 2006. Myostatin short interfering hairpin RNA gene transfer increases skeletal muscle mass. J Gene Med, 8(9): 1171-1181.
McCroskery S, Thomas M, Maxwell L, et al. 2003. Myostatin negatively regulates satellite cell activation and self-renewal. J Cell Biol, 162: 1135-1147.
McPherron A C, Lawler A M, Lee S J. 1997. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature, 387(6628): 83-90.
McPherron A C, Lee S J. 1997. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA, 94(23): 12457-12461.
Patel K, Amthor H. 2005. The function of myostatin and strategies of myostatin blockade-new hope for therapies aimed at promoting growth of skeletal muscle. Neuromuscul Disord, 2: 117-126.
Taylor W E, Bhasin S, Artaza J, et al. 2001. Myostatin inhibits cell proliferation and protein synthesis in C2C12 muscle cells. Am J Physiol Endocrinol Metab, 280(2): E221-E228.
Thomas M, Langley B, Berry C, et al. 2000. Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J Biol Chem, 275(51): 40235-40243.
Whittemore L A, Song K, Li X. 2003. Inhibition of myostatin in adult mice increases skeletal muscle mass and strength. Biochem Biophys Res Commun, 4: 965-971.
Wolfman N M, McPherron A C, Pappano W N. 2003. Activation of latent myostatin by the BMP-1/tolloid family of metalloproteinases. Proc Natl Acad Sci USA, 26: 15842-15846.
Xia X G, Zhou H, Xu Z. 2006. Department of biochemistry and molecular pharmacology. Biotechniques, 41(1): 64-68.
Yang J, Ratovitski T, Brady J P, et al. 2001. Expression of myostatin prodomain results in muscular transgenic mice. Mol Reprod Dev, 60: 351-361.
Zhu X, Hadhazy M, Wehling M, et al. 2000. Dominant negative myostatin produces hypertrophy without hyperplasia in muscle. FEBS Lett, 474(1): 71-75.
Q291
A
1006-8104(2014)-01-0031-08
Received 18 March 2013
Supported by the Major Special Projects of New Product Training of Transgenic Organisms (zx080072008-2008)
Feng Lin-he (1986- ), male, Master, engaged in the research of cell biology. E-mail: laohe31@126.com
* Corresponding author. Yan Yun-qin, professor, supervisor of Ph. D student, engaged in the research of cell biology. E-mail: yanyunqin@souhu.com
杂志排行
Journal of Northeast Agricultural University(English Edition)的其它文章
- Study on Relationship Between Cucumber Germplasm and Propamocarb Residue Using Subjective Rating Technique
- Cloning and Expression Analysis of Mlo Gene from Pericallis hybrida B. Nord.
- Simulation of in situ Root Decomposition of Two Barley Cultivars
- Vertical Migrating and Cluster Analysis of Soil Mesofauna at Dongying Halophytes Garden in Yellow River Delta
- Application of Protein Feed Processed by Microbial Fermentation to Dairy Cow
- Cloning and Sequence Analysis of Y-box Binding Protein Gene in Min Pig
