Cloning and Expression Analysis of Mlo Gene from Pericallis hybrida B. Nord.
2014-03-07WangJingangLiWeiRenHongweiLvYuandaBaiDingandXuJing
Wang Jin-gang, Li Wei, Ren Hong-wei, Lv Yuan-da, Bai Ding, and Xu Jing
College of Horticulture, Northeast Agricultural University, Harbin 150030, China
Cloning and Expression Analysis of Mlo Gene from Pericallis hybrida B. Nord.
Wang Jin-gang*, Li Wei, Ren Hong-wei, Lv Yuan-da, Bai Ding, and Xu Jing
College of Horticulture, Northeast Agricultural University, Harbin 150030, China
The full-length Mlo gene was obtained by reverse transcription polymerase chain reaction (RT-PCR) and RACE. The result of sequence analysis indicated that Mlo gene from Pericallis hybrida B. Nord. contained about 1 296 bp open reading frame and encoded 431 amino acids. According to the comparison of the exogenous gene sequences by BLAST analysis and phylogenetic analysis, Mlo gene shared over 85% nucleotide homology and 98% amino acid homology. Finally, through semi-quantitative-PCR and fluorescence quantitative analysis, we found that Mlo gene showed the highest expression levels in leaves and the lowest in roots after inoculated with powdery mildew pathogen for different days.
Pericallis hybrida B. Nord., Mlo gene, cloning, sequence expression analysis
Introduction
Pericallis hybrida B. Nord. is a perennial herb of the Asteraceae. It is an important ornamental potted flower in the New Year's Day and the Spring Festival. Study on Pericallis hybrida B. Nord has a broad prospect and practical significance. In recent years, powdery mildew has gradually become a serious disease in Cineraria, and constraints the commercialization industry development of Cineraria. Traditional biological control methods will cause the pathogen resistance and environmental pollution problems. Therefore, the use of genetic engineering has important significance to nurture Cineraria germplasm resources with broadspectrum resistance to powdery mildew (Liu et al., 2010).
The prevention mechanism of Mlo gene of powdery mildew is not very clear. But studies on tomatoes and other crops had shown that Mlo gene caused the plant to produce spontaneous leaf cell death phenomenon and played a negative regulatory factor role in the regulation of resistance (Pryor et al., 2000; Bonnema et al., 2003). The durable resistance to non-pathogenic small species of Mlo gene might be involved in the regulation process of powdery mildew (Freialdenhoven et al., 1997).
Therefore, the cloning of Mlo gene as well as the study of its structure and function provided new ideas for the breeding of disease resistance. However, the study of Mlo gene of Pericallis hybrida B. Nord. had not yet reported. In this study, we took Pericallis hybrida B. Nord. as the material, the full-length Mlo gene was cloned by RT-PCR and RACE methods. The protein structure, evolution status and expression pattern of different tissues and biological stresses werepredicted and analyzed by bioinformatics methods.
Materials and Methods
Plant materials and reagents
Pericallis hybrida B. Nord. was provided by Horticulture Experimental Station of Northeast Agricultural University. The roots, stems and leaves were quickfrozen in liquid nitrogen and stored at –80℃ refrigerator.
Total RNA extraction kit was bought from Trans-Gene Biotech. 3'-full RACE kit, 5'-full RACE kit, TaKaRa LA Taq, DL2000 and restriction enzymes were purchased from TaKaRa. Primers designing, gene synthesis and gene sequencing were carried out in Boshi Biotech.
Full-length cloning of Mlo gene of Pericallis hybrida
Compared the nucleotide and amino acid sequences of the exogenous Mlo gene with GenBank, selected the appropriate conservative area to design degenerate primers by Primer5.0 (Panstruga et al., 1997).
Forward: 5'-CAYCARCTGCAYATHTTCAT-3'
Reverse: 5'-GGVAGAGTCACATAGCTGCA-3'.
The first-strand cDNA of healthy leaves was taken as the template for PCR amplification. PCR amplification program: 94℃ 3 min for pre-denaturation (94℃for 30 s, 51℃ for 1 min, 72℃ for 1 min), 30 cycles, 72℃ 10 min for extension. The swimming with expected size (800 bp) was collected with DNA Gel Extraction Kit. The product was connected to pMD18 T-vector and transformed into DH5α. The positive clones were sequenced by blue and white screening and plasmid digested in Boshi Biotech Co., Ltd.
Specific primers were designed according to the middle fragment sequence, 5' primer sequences: outer: 5'-CATGGCTACATGCTGACAGCCTA-3', inner: 5'-CGCGGATCCACAGCCTACTGATGATCAGT CGATG-3'; 3' primer sequences: outer: 5'-TACCGT CGTTCCACTAGTGATTT-3'. Rapid amplification of cDNA was carried out in accordance with RACE kit instructions. Gel extraction, product cloning and sequencing were described above.
Protein sequence alignment and phylogenetic analysis
The amino acid sequences encoded by cDNA sequence and open reading frame were contrasted by BLAST analysis in GenBank database. The obtained Mlo gene sequences were alignment with Mlo homologous protein sequence landing in GenBank by CLASTALW2 software. The phylogenetic tree was constructed by DNAMAN (Freialdenhoven et al., 1997).
Expression analysis of Mlo gene
The expression levels of Mlo gene in three tissues of Pericallis hybrida B. Nord. were analyzed by semi-quantitative. The expression differences after inoculated with powdery mildew pathogen for 0, 1, 3, 5 and 7 days were analyzed by real-time fluorescence quantitative (Dechert et al., 2003). Actin internal reference primers were designed based on Actin gene EST sequences of Pericallis hybrida B. Nord. in GenBank. Special primers as follows:
Mlo gene:
Forward: 5'-CAAGATGAGGAAGTGGAAGG-3'
Reverse: 5'-AACTTTAGCAACTGAGGTGA-3'.
Actin gene:
Forward: 5'-TCAACCCCAAAGCTAACAG-3'
Reverse: 5'-ATCCTCCGATCCAGACACT-3'
PCR amplification program: 94℃ 3 min for predenaturation (94℃ for 30 s, 55℃ for 45 s, 72℃ for 1 min), 30 cycles, 72℃ 10 min for extension. Amplification products were purified by 1% agarose gel electrophoresis and gel imaging system. After the reaction, the kinetics and melting curve analysis charts were obtained from IQ5 Detection System by iCycler software.
Results
Cloning of ORF full-length cDNA of Mlo gene
It could be seen that full-length cDNA contained a1 296 bp open reading frame which including a 586 bp 5'-UTR and a 372 bp 3'-UTR (Figs. 1-4).
Homology analysis of Mlo gene encoding amino acid sequences
The results of sequence comparison by CLASTALW2 showed that it shared over 75%-85% nucleotide homology and 85%-100% amino acid homology,where was completely homologous with Arabidopsis thaliana (Fig. 5).
For preliminary analysis of the homology relationship of Mlo gene with other plants, we constructed phylogenetic tree. The overall analyses showed that the genetic distances of Mlo gene among these plants were very small, so Mlo gene was conserved (Fig. 6).

Fig. 1 Electrophotogram of RT-PCR amplification by Mlo gene
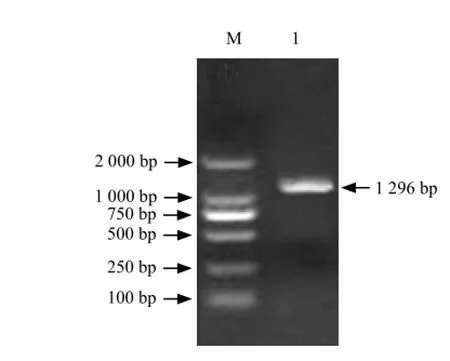
Fig. 2 RT-PCR amplification of full-length cDNA
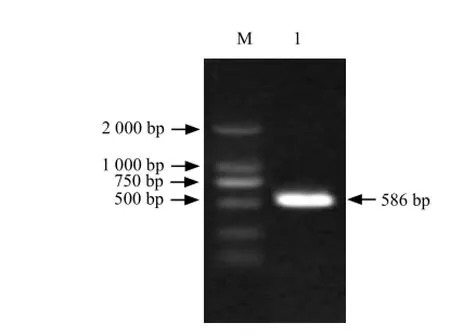
Fig. 3 RT-PCR amplification of 5′-RACE
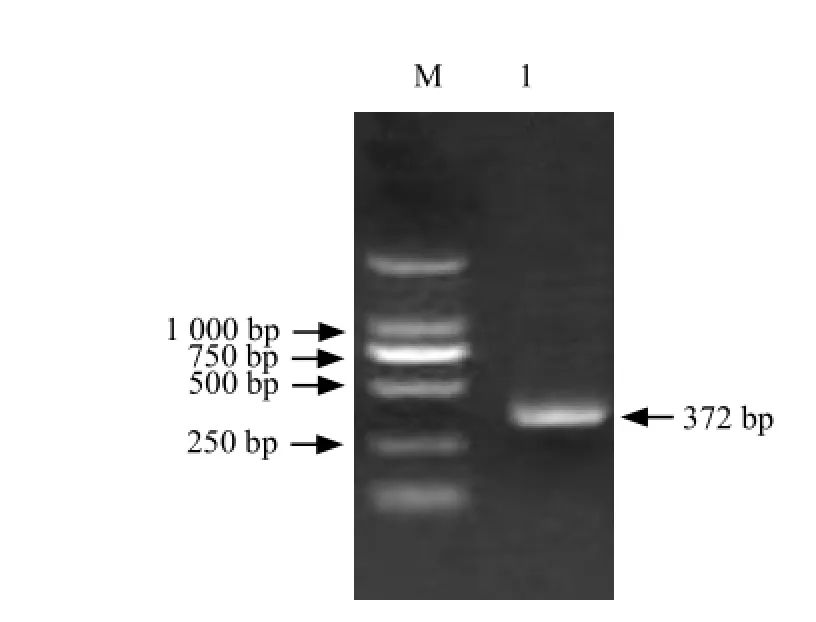
Fig. 4 RT-PCR amplification of 3′-RACE
Expression analysis of Mlo gene
The expression quantity of Mlo gene in roots, stems and leaves was different. The infection time also influence the expression quantity of Mlo gene. The expression levels of leaves were the highest, and the lowest were roots (Fig. 7).
The expression of Mlo gene started on 0 day. The expression levels were significantly enhanced on the 1st day, and then they reduced, but still higher than that without induced. The expression levels began to decline until the 7th day (Fig. 8).
Through the fluorescence quantitative analysis of Mlo gene from different tissues of Pericallis hybrida B. Nord. and inoculated with powdery mildew pathogen for different days, the results showed thatthe leaves had high relative expression abundance, and the lowest were roots (Fig. 9). The highest relative expression abundances were on the 1st day and the lowest were on the 7th day (Fig. 10).

Fig. 5 Multiple alignment of nucleic acid sequences of Mlo gene

Fig. 6 Dendrogram of deduced amino acid sequence of Mlo gene from some plants
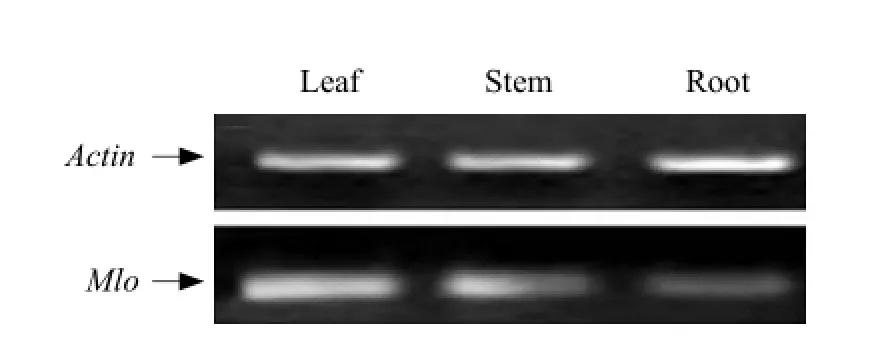
Fig. 7 RT-PCR analysis of Mlo gene expression in root, stem and leaf

Fig. 8 Mlo gene expression induced with powdery mildew for different times

Fig. 9 Real-time PCR analysis Mlo gene expression in root, stem and leaf
Discussion
In this study we took Pericallis hybrida B. Nord. as the material, the full-length of Mlo gene was cloned by RT-PCR and RACE. Homology and bioinformatics analysis results showed that Mlo gene was conserved, nucleotide homology was over 80%, and the amino acid sequence homology was over 90%, which could be used as a kind of candidate genes of plant evolutionary studies (Dietz et al., 1995; Schaefer, 1995).
Mlo gene had tissue-specific expression patterns (Aki et al., 1999). According to the relevant literature, RmMlo genes were expressed in leaves, petals and flowers of Rosa multiflora, but not in roots (Qiu et al., 2011). However, all of them were expressed in Pericallis hybrida. The leaves had the highest expression levels, and the lowest were roots. The differences were probably, due to the functional differentiation of dicotyledon taxa plants and resistance evolution-control mode of woody and herbaceous plants.

Fig. 10 Real-time PCR analysis Mlo gene expression induced with powdery mildew for different times
Powdery mildew-induced also had some impacts on expression of Mlo gene. Through the fluorescence quantitative analysis of Mlo gene from different tissues of Pericallis hybrida B. Nord. and inoculated with powdery mildew pathogen for different days, the results showed that the leaves had the highest relative expression abundance, and the lowest were roots. The highest relative expression abundances of Mlo gene were on the 1st day, and the lowest were on the 7th day. The other days they increased slightly. These were similar to the expression patterns of reported Arabidopsis thaliana and Cucumis melo. Mlo gene might be involved in the regulation process of powdery mildew, but its regulatory mechanism needed further study.
Conclusions
In this study, we have successfully cloned the fulllength sequence of Mlo gene nucleotide sequence of Pericallis hybrida B. Nord. by RT-PCR and RACE. The full-length Mlo gene contained a 1 296 bp open reading frame and encoded 431 amino acids. The results of homology analysis and bioinformatics analysis showed that it had higher similarity with the dicotyledonous plants and the monocotyledonous were relatively low. Through semi-quantitative-PCR and fluorescence quantitative analysis, we found that Mlo gene had tissue-specific expression patterns. For further study of the structure and function of Mlo gene, we would build the gene expression vector and silence gene carrier, and then transform into the high resistance and high sense of Pericallis hybrida material to verify its biological function and lay the foundation for resistance gene engineering breeding.
References
Aki A, Kazuaki I, Takeshi T T, et al. 1999. Quantitation of hepatitis Bviru genomic DNA by real-time detection PCR. Joumal of Clinieal Mierobiology, 37(9): 2899-2903.
Bonnema G, Bai Y, Huang C C, et al. 2003. QTLs for tomato powdery mildew resistance (Oidium lycopersice) in Lycopersicon parviflorum G1.160 co-localize with two qualitative powdery mildew resistance genes. Mol Plant-Microbe Interact, 16(7): 169-176.
Dechert C, Schultheiss H, Kogel K, et al. 2003. Functional analysis of barley RAC/ROP G-protein family members in susceptibility to the powdery mildew fungus. The Plant Journal, 36(4): 589-601.
Dietz K J, Rudloff S, Ageorges A, et al. 1995. Subunit E of the vacuolar H+-ATPase of Hordeum vulgare L: cDNA cloning, expression and immunological analysis. The Plant Journa, 18(5): 521-529.
Freialdenhoven A, Peterhansel C, Kurth J, et al. 1997. Interaction analyses of genes required for resistance responses to powdery mildew in barley reveal distinct pathways leading to leaf cell death. The Plant Cell Online, 9(6): 1397-1409.
Liu J C, Zhang Y Y, Li X Y. 2010. The prevention of major pests and diseases of Pericallis hybrida. Modern Gardening, 2(4): 43-44.
Pryor T, Ellis J, Dodds P. 2000. Structure, function and evolution of plant disease resistance genes. Current Opinion in Plant Biology, 3(5): 278-284.
Panstruga R, Bueschges R, Hollricher K, et al. 1997. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell, 88(3): 695-705.
Schaefer B C. 1995. Resolution in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal Biochemical, 227(1): 255-273.
Qiu X X, Bao M Z, Wang Q G, et al. 2011. The cloning and expression analysis of powdery mildew resistance gene RmMlo of Rosa multiflora Thunb. Modern Gardening, 38(10): 1999-2004.
Q943
A
1006-8104(2014)-01-0010-06
Received 22 March 2013
Supported by Postdoctoral Scientific Research Foundation of Heilongjiang Province (LBH-Q10144); the Natural Science Foundation of Heilongjiang Province (C201112); Northeast Agricultural University Doctoral Research Fund (200830)
Wang Jin-gang (1974-), male, professor, engaged in the research of genetic breeding of ornamental plants and biotechnology. E-mail: Wangjingang99@yahoo.com.cn
杂志排行
Journal of Northeast Agricultural University(English Edition)的其它文章
- Study on Relationship Between Cucumber Germplasm and Propamocarb Residue Using Subjective Rating Technique
- Simulation of in situ Root Decomposition of Two Barley Cultivars
- Vertical Migrating and Cluster Analysis of Soil Mesofauna at Dongying Halophytes Garden in Yellow River Delta
- Effect of MSTN Propeptide and shRNA Co-expression Vector on Proliferation of Skeletal Muscle Satellite Cells
- Application of Protein Feed Processed by Microbial Fermentation to Dairy Cow
- Cloning and Sequence Analysis of Y-box Binding Protein Gene in Min Pig
