毕赤酵母中植酸酶和内切葡聚糖酶共表达菌株的构建及其高效表达
2010-09-29吴振芳唐自钟陈惠韩学易赖欣吴琦
吴振芳,唐自钟,陈惠,韩学易,赖欣,吴琦
四川农业大学生命科学与理学院,雅安 625014
毕赤酵母中植酸酶和内切葡聚糖酶共表达菌株的构建及其高效表达
吴振芳,唐自钟,陈惠,韩学易,赖欣,吴琦
四川农业大学生命科学与理学院,雅安 625014
植酸酶和内切葡萄糖苷酶广泛应用于动物饲料添加剂中,能更有效地帮助饲料的利用,同时为动物的生命活动提供必需的重要物质。首先构建重组质粒pPICZα-EG,经过线性化后,电转化到含有植酸酶phyA基因的毕赤酵母感受态细胞中,构成含有植酸酶基因和内切葡萄糖苷酶基因的重组体菌株 GS115-phyA-EG。经甲醇诱导后其活性分别达到出发菌株GS115-phyA和GS115-EG的39.4%和56.2%。酶学性质的分析显示,最适反应温度和最适反应的pH值分别为55℃和5.5,植酸酶和内切葡萄糖苷酶在温度为45~55℃℃,pH值为4.5~5.5时,酶活相对稳定,均能达到最高酶活的80%以上。结果表明这种在同一个系统种同时表达2种酶的方法能更好地节约时间和成本,使其在饲料工业的应用上有更广阔的前景。
植酸酶,内切葡聚糖酶,毕赤酵母,混合表达
Abstract:Both phytase and endoglucanase are additives in feed for mono-gastric animal known for their effects. Recombinant vector pPICZα-EG was constructed and transformed to GS115-phyA, a Pichia pastoris strain that had integrated with phytase gene,generating GS115-phyA-EG. Both phytase and endoglucanase activities in the supernatant were determined after methanol induction of GS115-phyA-EG. Phytase and endoglucanase activity reached 39.4% and 56.2% activity compared to GS115-phyA and GS115-EG,respectively. Properties of the mixed enzyme suggest that the optimal temperature and pH value be 55°C and 5.5 respectively. Both phytase and endoglucanase showed greater than 80% activity across temperature ranges 45°C to 55°C and pH ranges 4.5 to 5.5.Expressing more than one enzyme in one system could save time and money during induced expression, and the mixed enzyme might apply for treating forge before feeding with poultry.
Keywords:phytase, endoglucanase, Pichia pastoris, mix-expression
Introduction
Phytases (myo-inositol hexakisphosphate phosphohydrolase;EC 3.1.3.8) belong to the family of histidine acid phosphatases and are ubiquitous in plants and fungi.They catalyze phosphomonoester cleavage of phyticacid (myo-inositol hexakisphosphate), which is the major storage form of phosphorus in plants[1].Supplementing with inorganic phosphate to feed can solve the problem that non-ruminants such as pigs and poultry, virtually lack phytase activity in their digestive tract. However, this supplimentation imposes ecological problems. Since the application of the phytic acid has been available in animal feeds, phytase been a good choice to take place of inorganic phosphate, which has solved ecological problems to some extent[2]. Cellulase is a term for a group of enzymes able to transform cellulose to glucose. This group mainly consists of endo-1,4-β-glucanase (EC 3.2.1.4), 1,4-β-D-cellobiohydrolase (EC 3.2.1.9) and 1,4-β-glucosidase (EC 3.2.1.21). These three enzymes cooperate to translate cellulose to glucose completely.The major sources of these enzymes are microorganisms, including bacteria and fungi,especially those that utilize cellulose as their energy source[3-4]. Endoglucanases, especially from fungi,have drawn the most interest in the previous cellulase study[5]. Recently, bacterial cellulases have taken on satisfactory application performance and economic value, because they show relative stability in neutral and basic conditions[6].
Both phytases and endoglucanases are additives in forage for mono-gastric animal, which acknowledged for their effects[7]. However, most of enzymes may be digested by proteinases in vivo, which reduced their effectiveness as additives. Therefore, treating forage with mixed enzymes in vitro would be more effective for feeding domestic animals. Expressing more than one enzyme in a suitable expression system simultaneously became available for obtaining mixed enzymes.
Pichia pastoris is a methylotrophic yeast,well-known for its ability to express high levels of heterologous proteins[8-9]. We have constructed a recombinant Pichia pastoris strain, capable of overexpressing phytase and endoglucanase simultaneously.The properties of phytase and endoglucanase expressed in this system have also been discussed.
1 Materials and methods
1.1 Experimental materials
Escherichia coli DH5α was used as host for DNA manipulation and transformation. DH5α which contains endolucanase gene was constructed in our laboratory[10].Plasmid pPICZα was used for endoglucanase expressing vector construction. Pichia pastoris GS115 and GS115-phyAwhich contains phyA gene, were used as host for electroporation and expression. These strains were constructed and stored in our laboratory[11]. Pfu DNA polymerase (TIAGEN Beijing, China) and the restriction enzymes (TaKaRa Dalian, China) used in this study were commercially available; and the primer pair was synthesized by Invitrogen (Shanghai, China).
1.2 Recombinant vector construction for endoglucanase expression
Amplification of endoglucanase gene was carried out by Pfu DNA polymerase, and a pair of primers were designed and synthesized. The forward and reverse primers were 5′-CCGGAATTC GCAGAGAC AAAAACGCCAGT-3′ and 5′-GAATGCGGCCGC CT AATTTGGTTCTGTTC-3′; the underline indicated EcoR I and Not I site. The PCR products were digested by EcoR I and Not I restriction enzymes then ligated into pPICZα at the corresponding restriction sites, to generate pPICZα-END. The recombinant was identified by restriction analysis and DNA sequencing.
1.3 Transformation of P. pastoris GS115 and GS115- phyA with pPICZα-END
An amount of 10 μg pPICZα-END plasmid was linearized with Sac I, and then electroporated into P. pastoris GS115 and GS115-phyA containing phyA gene. The transformants were plated on YPD-Zeocin agar plates (1% yeast extract, 2% peptone, 2% glucose and 100 μg/mL zeocin), and incubated at 30°C for 2−4 days until colonies obtained. GS115-phyA which integrated with pPIC9k-phyA plasmid didn’t grow in YPD-Zeocin agar plates. Only the colonies inserted with pPICZα-END were able to grow on this culture.The recombinant P. pastoris GS115 strains were named GS115-END and GS115-phyA-END, respectively(Fig. 1).
1.4 Expression of GS115-END and GS115-phyAEND in shaking flask
The colonies were selected randomly and inoculated into 3 mL BMGY medium (2% peptone, 1% yeast extract, 100 mmol/L potassium phosphate (pH 6.0),1.34% YNB, 1% glycerol and 4×10−5% biotin) in 50 mL flasks. After shaken at 30°C for 18–22 h, the cells were collected by centrifugation (4 000 r/min,10 min), then resuspended in 5 mL BMMY medium(BMMY with 0.5% methanol instead of 1% glycerol in BMGY). The culture was shaken for 48 h at 30°C before measuring the enzymatic activity in supernatant.100% methanol was supplemented every 24 h to a final concentration of 1% for maintaining induction throughout the induction phase. GS115-phyA,GS115-END and GS115-phyA-END were expressed.Each strain was inoculated into 25 mL BMGY medium in 250 mL flasks. The cells were collected until the OD600reached 2−6, then resuspended with 100−200 mL BMMY medium (the final cell density OD600=1) in 1 000 mL flasks[12]. GS115-phyA and GS115-END were also cultured and expressed together for mix-expression of phytase and endoglucanase to compare with the capability of GS115-phyA-END.After expressed for 4 d at 30°C, the supernatants were used for determination of enzymatic activity and enzymatic characters.

Fig. 1 Schematic diagram of construction of GS115-phyA-End.
1.5 Enzymatic activity assays
The activities of phytase and endoglucanase were determined in the culture supernatants. The phytase activity was estimated by determining the freephosphorus concentration and following the method described by Han et al[13]. The crude enzyme was diluted with acetic acid buffer (pH 4.6), and 2 mL diluted enzyme was transferred to a 10 mL tube. Then,4 mL sodium phytate (diluted with acetic acid buffer)was added to start the reaction. The reaction was stopped by adding 4 mL color/termination solution after incubating at 37°C for 30 min. Blanks were enzyme with color/termination solution before adding sodium phytate.
The endoglucanase activity was determined using classical method described by Mandels et al[14]. In this method CMC-Na was used as substrate; the amount of reduced sugars which released from the assay was detected by the DNS method. 200 μL supernatant was diluted to 1 mL PBS buffer (pH 6.0). 100 μL diluted enzyme and 1 mL 1% CMC-Na were mixed in a 10 mL tube and incubated at 50°C for 30 min. The reaction was terminated adding 2.5 mL DNS, then boiled for 5 min. Blanks were measurement of the endoglucanase activity before adding CMC-Na.
1.6 Enzyme characterization assays
The optimal temperature for reaction was determined at various temperatures from 30°C to 85°C in corresponding buffer. To determine thermostability,the enzyme was pre-incubated at 40°C to 85°C in buffer for 30 min, then perform the standard activity assay. The enzyme and substrate were diluted to pH range from 3.0 to 10.0 using various buffers to estimate the optimal pH. Also the enzyme was pre-incubated at 50°C with different buffers which has the pH range from 3.0 to 10 to determine the pH stability. The standard DNS activity assay was then performed.
1.7 Genome isolation and amplification of genomic DNA
Chromosome DNA isolation and genome PCR were performed in order to check the integrity of phytase and endoglucanase gene in Pichia genome DNA.Pichia genomic isolation was performed following the method described by Holm et al[15]. Genome PCR was completed through a standard PCR amending template with Pichia genome DNA. The primers used for amplifying phytase[16]and endoglucanase genes were added respectively to the reaction.
1.8 The genetic stability of the recombinant strain
The recombinant colonies were selected and inoculated into 25 mL BMGY medium and incubated at 30°C in a shaking incubator until A600of the culture medium reaches 2–6. The cells were centrifuged at 4 000 r/min for 10 min then resuspended in 100 mL BMMY medium. During the 108 h incubation period,methanol was added every 24 h to reach a final volume fraction of 1%. Then the enzyme activity was determined and 1ml cells were used for the next inducible expression. And this experiment was performed for 10 generations of the recombinant strain.
2 Results
2.1 Expression of endoglucanase and phytase enzyme
The recombinant vector pPICZα-END was constructed and identified by restriction analysis and DNA sequencing. Then, pPICZα-END plasmid was linearized with Sac I, and electroporated into P. pastoris GS115 and GS115-phyA. Ten colonies from each strain were randomly selected and methanol inductions continued for 48 h. Crude enzyme in supernatant wasmeasured through standard activity assay. Only two colonies of GS115-phyA-END had activity of both phytase and endoglucanase; the rest colonies had either phytase activity or endoglucanase activity (Table 1).
The 4th and 8th colonies showed both activity of phytase and endoglucanase, however, they lost both enzymatic activities to some extent. The phytase activity of these two colonies was only about half of it in other strains. Meanwhile, endoglucanase activity reached to approximately 80%. The 6th and 4th in GS115-END and GS115-phyA-END were selected for the following experiments.
The GS115-phyA, GS115-END and GS115-phyAEND were performed expression in scale, the supernatants at 0, 12, 24, 36, 48, 60, 72, 84, 96, 108 and 120 h were sampled for enzymatic activity determination (Fig. 2).
Supernatants at 96−108 h reached the maximal activity. GS115-phyA-END showed 13 726.7 U/mL phytase activity and 364.5 U/mL endoglucanase activity, 39.4% of GS115-phyA (34 820.6 U/mL) and 56.2% of GS115-END (649.0 U/mL). In the mixexpression flask, both the activities of phytase and endoglucanase are also reduced, which suggested the total expressing mass of heterogolous proteins in P. pastoris may have a limitation.
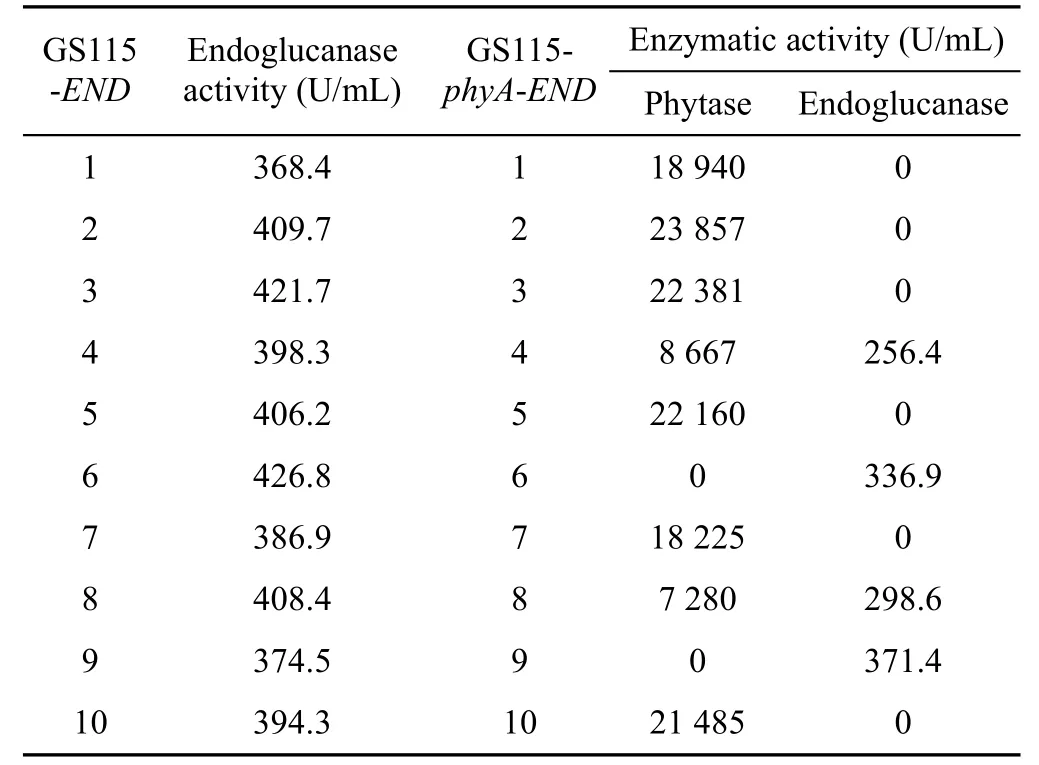
Table 1 Enzymatic activity screening of GS115-END and GS115-phyA-END

Fig. 2 Phytase and endoglucanase activity determination of P. pastoris recombinant strains. (A) Phytase activity of GS115-phyA. (B)Endoglucanase activity of GS115-END. (C) Phytase and endoglucanase activities of GS115-phyA-END. (D) Phytase and endoglucanase activities of mix-expression of GS115-phyA and GS115-END.
2.2 Enzymatic characters assays
Phytase and endoglucanase mixed enzyme properties were determined using crude enzyme expressed in GS11-phyA-END. The optimal temperatures of phytase and endoglucanase were 55°C and 65°C, respectively.The crude enzyme exhibited above 60% both phytase and endoglucanase activities across narrow temperature(35°C−55°C) ranges (Fig. 3).
Both of their activity lost much after being treated at 70°C for 30 min (Fig. 4).
The optimal pH values of the two enzymes were 4.5 and 6.0, respectively. The pH ranges where both phytase and endoglucanase can reach above of 60% of their highest activities were between 4.0−6.0 (Fig. 5).
The phytase was stable at acid conditions (pH 3.0−5.5), while the endoglucanase can preserve most of activity at neutral and basic conditions (even when pH value reached 10.0) (Fig. 6).
2.3 Pichia genomic isolation and genome PCR
Chromosome DNA isolation and genome PCR were performed to verify the integration of phyA gene and endoglucanase gene in Pichia genome DNA. Genome DNAs of GS115-phyA, GS115-END and GS115-phyAEND were then used as templates for amplifying phytase and endoglucanase gene (Fig. 7).
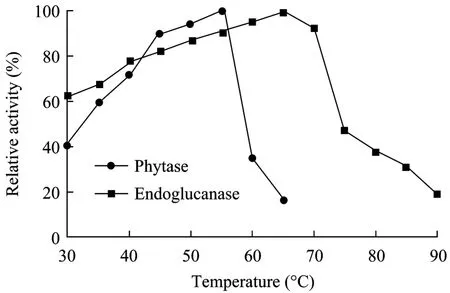
Fig. 3 Optimal temperature of phytase and endoglucanase.

Fig. 4 Thermostability of phytase and endoglucanase.
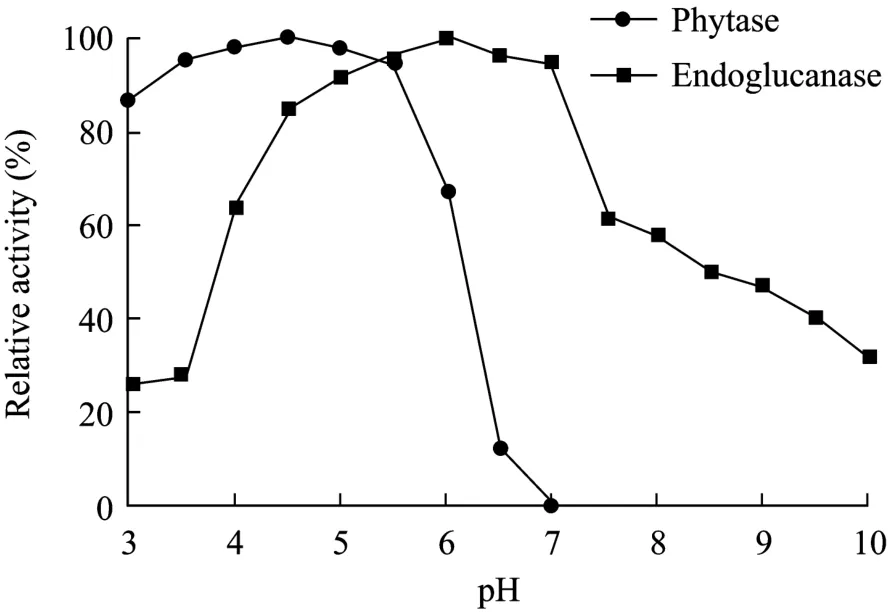
Fig. 5 Optimal pH profile of phytase and endoglucanase.
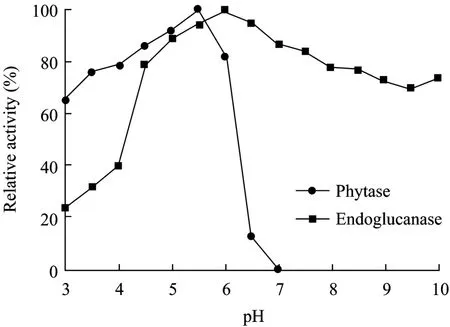
Fig. 6 pH stability of phytase and endoglucanase.

Fig. 7 Genome PCR of GS115-phyA, GS115-END and GS115-phyA-END. M: DNA marker VII; E and P indicate genome PCR were performed adding the primers amplifying endoglucanase and phytase gene, GS115-phyA genome DNA can be only used for amplifying phyA gene fragment, and it is contrary to GS115-END. However, applying GS115-phyA-END genome DNA as template, we only obtained phyA and endoglucanase gene fragments in accordance to the corresponding primers.The results indicated that both phytase and endoglucanase gene have integrated into the GS115-phyA-END chromosome DNA.
2.4 The genetic stability of the recombinant strain
The results of 10 generations of consecutive culture and the activity determination indicated that the recombinant strain GS115-phyA-END remained stable on the growth and the expression of phytase and endoglucanase. This result showed that the recombinant strain is genetic stable (Table 2).
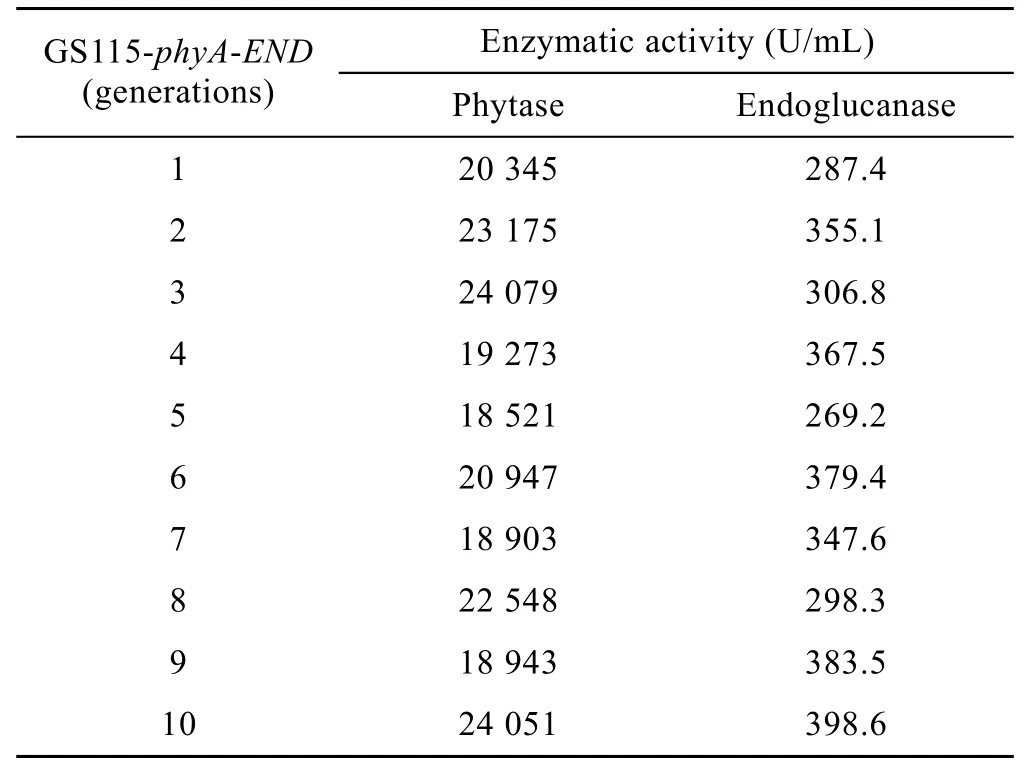
Table 2 Generations and enzymatic activity of GS115-phyA-END
3 Discussion
Pichia pastoris is methylotrophic yeast. It is well-known for its ability to express high levels of heterologous proteins. Many heterologous proteins have been reported over-expression through this system[8-9,17]. Aspergillus Niger N25 phytase gene(phyA) has been expressed in Pichia pastoris in our previous research, and the phytase activity reached above 30 000 U/mL which is more than 50 fold compared with the original strain[16]. Many factors,such as integration form, integration sites and multicopy inserts, can affect the heterologous proteins expressing level in Pichia pastoris[18]. Multiple copy insertion in Pichia has been reported to improve the expressing level in some cases, and some expressing vectors, such as pPIC9k and pAO815, have been designed for multicopy insertion in vivo and in vitro respectively[19-20]. Integration form has also been demonstrated as an important factor in affecting the expressing level. Heterologous fragments can be inserted into Pichia pastoris chromosome DNA at AOX1 and HIS4 region according to the linearization sites of expressing vector[8].
Both phytase and endoglucanase are additives in forage for mono-gastric animal, which are often pelleted, and then added to animal feeds. However,many natural enzymes can’t withstand the temperatures during the pelleting process[21]. In the other hand,most of enzymes may be digested by proteinase in vivo,which reduced much of their effect as additives[7].Treating forage with mixed enzymes in vitro was considered more effective for feeding domestic animal, and more than one enzyme expressed in Pichia pastoris can be suitably developed for obtaining mixed enzyme expediently.
In this study, endoglucanase gene has integrated into GS115-phyA genome DNA containing phytase gene.Activity screening was performed to obtain recombinant Pichia containing both phytase and endoglucanase gene. This recombinant strain was named GS115-phyA-END. Some strains showed either phytase or endoglucanse activity, which demonstrated several integration form happened, such as insertion and replacement. In our previous study, pPIC9k-phyA was linearize with BspE I then transformated into GS115. Therefore the integration site located at HIS4 region[10]. In this study, pPICZα-END was inserted into AOX1 region of Pichia chromosome DNA.Genome PCR of GS115-phyA, GS115-END and GS115-phyA-END was performed to verify the insertion, and the results indicated that both phytase and endoglucanase gene had inserted into the GS115-phyA-END genome DNA. Phytase and endoglucanase activities expressed in GS115-phyAEND were 13 726.7 U/mL and 364.5 U/mL, 39.4% of GS115-phyA (34 820.6 U/mL) and 56.2% of GS115-END (649.0 U/mL), respectively. GS115-phyA and GS115-EG were cultured and expressed in a flask for controlling the two enzymes expression level of GS115-phyA-END. The results showed that both the two enzymatic activities also reduced above the half of the highest activities. The total expression mass of heterologous proteins in Pichia may have a limitation,especially with a constant capacity. However, inserting two genes into one strain provided convenience for strain store, expression, and so on. Some colonies which resistant to zeocin, showed low endoglucanase activity(about 80 U/mL). The cause may be the genetic stability of transformant[22], which will be discussed in the future study.
The enzymatic properties of phytase and endoglucanase expressed in GS115-phyA-END were determined in this study. Both of the enzymes were demonstrated same as expressed alone. The crude enzyme exhibited more than 60% both phytase and endoglucanase activities between temperature range35°C to 55°C, and the optimal temperature of the mixed enzyme was estimated to be 55°C. The pH values where both phytase and endoglucanase can reach above 60% of their highest activities were located at pH 4.0−6.0 range, and pH 5.5 was considered as the optimal pH. Expressing more than one relative enzyme in one system offered convenience for obtaining these enzymes simultaneously, which can save time and money for induced expression. In the other hand, since it was reported forge additives would reduce by digestion of proteinase in vivo, treating forge with mixed enzyme before feeding with animal was considered as more effective strategy. These enzymatic properties could be convenient for application of the mixed enzyme to industry.
REFERENCES
[1] Wodzinski RJ, Ullah AH. Phytase. Adv Appl Microbiol,1996, 42: 263−302.
[2] Mullaney EJ, Daly CB. Advances in phytase research. Adv Appl Microbiol, 2000, 47: 157−199.
[3] Marchessault RH, Sundararajan PR. The polysaccharides//Aspinall GO, ed. Cellulose. New York: Academic Press,1993: 11−95.
[4] Lee RL, Paul JW, Willem HZ, et al. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev, 2002, 66(3): 506−577.
[5] Monika S, Kubicek CP. Regulation of Trichoderma cellulase formation: lessons in molecular biology from an industrial fungus. Acta Microbiol Immunol Hungar, 2003,50(2/3): 125−145.
[6] Bequin P, Millet J, Chauvaux S, et al. Bacterial cellulases.Biochem Soc Trans, 1992, 20(1): 42−46.
[7] Reddy VA, Venu K, Rao DE, et al. Chimeric gene construct coding for bio-functional enzyme endowed with endoglucanase and phytase activities. Arch Microbiol,2009, 191(2): 171−175.
[8] Cregg JM, Vedvick TS, Raschke WC. Recent advances in the expression of foreign genes in Pichia pastoris. Nat Biotechnol, 1993, 11: 905−910.
[9] Romanos M, Wellcome G, UK B. Advances in the use of Pichia pastories for high-level gene expression. Curr Opin Biotechnol, 1995, 6: 527−533.
[10] Chen H, Xu B, Liao JH, et al. Expression of endoglucanase gene in Bacillus megaterium and characterization of recombinant enzyme. Hereditas, 2008,30(5): 649−654.
[11] Chen H, Zhao HX, Wang HN, et al. Increasing expression level of phytase gene (phyA) in Pichia pastoris by changing rare codons. Chin J Biochem Mol Biol, 2005,21(2): 171−175.
陈惠, 赵海霞, 王红宁, 等. 植酸酶基因中稀有密码子的改造提高其在毕赤酵母中的表达量. 中国生物化学与分子生物学报, 2005, 21(2): 171−175.
[12] Wu ZF, Chen H, Zeng M, et al. Over-expression study of Bacillus subtilis neutral endoglucanase gene in Pichia pastories. J Agri Biotechnol, 2009, 17(3): 529−535.
[13] Han YM, Wilson DB, Lei XG. Expression of an Aspergillus niger phytase gene (phyA) in Saccharomyces cerevisiae. Appl Environ Microbiol, 1999, 65(5):1915−1918.
[14] Mandels M, Andreotti R, Roche C. Measurement of saccharifying cellulose. Biotechnol Bioeng Symp, 1976, 6:21−33.
[15] Holm C, Meeks-Wagner DW, Fangman WL, et al. A rapid,efficient method for isolating DNA from yeast. Gene,1986, 42: 169−173.
[16] Wang HN, Ma MG, Wu Q, et al. Over-expression research of Aspergillus niger N25 phytase gene (phyA) in Pichia pastoris. Mycosystema, 2001, 20(4): 486−493.
[17] Macauley-Patrick S, Fazenda ML, McNeil B, et al.Heterologous protein production using the Pichia pastoris expression system. Yeast, 2005, 22(4): 249−270.
[18] Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev, 2000, 24(1): 45−66.
[19] Daly R, Hearn MT. Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J Mol Recoqnit, 2005, 18(2):119−138.
[20] Luo HY, Huang HQ, Bai YG, et al. Improving phytase expression by increasing the gene copy number of appA-m in Pichia pastoris. Chin J Biotech, 2006, 22(4): 528−533.
罗会颖, 黄火清, 柏映国, 等. 增加植酸酶基因 appA-m的拷贝提高其在巴斯德毕赤酵母的表达量. 生物工程学报, 2006, 22(4): 528−533.
[21] Lehmann M, Pasamontes L, Lassen SF, et al. The consensus concept for thermostability engineering of proteins. Biochim Biophys Acta, 2000, 1543: 408−415.
[22] Zhu T, Guo M, Sun C, et al. A systematical investigation on the genetic stability of multi-copy Pichia pastoris strains. Biotechnol Lett, 2009, 31(5): 679−684.
Construction of a Pichia pastoris recombinant strain capable of over-expressing phytase and endoglucanase
Zhenfang Wu, Zizhong Tang, Hui Chen, Xueyi Han, Xin Lai, and Qi Wu
College of Life and Physical Science, Sichuan Agricultural University, Ya’an 625014, China
