Loop-mediated isothermal amplification assay targeting dnaN gene for rapid detection of Streptococcus suis
2016-08-25QIUHongLIUZhijieZHENGHan
QIU Hong, LIU Zhi-jie, ZHENG Han
(1. Hunan University of Medicine, Huaihua 418000,China;2. The 2nd Xiangya Hospital,Central South University, Changsha 410000,China;3. National Institute for Communicable Disease Control and Prevention,Chinese Center for Disease Control and Prevention, Beijing 102206, China)
Loop-mediated isothermal amplification assay targeting dnaN gene for rapid detection of Streptococcus suis
QIU Hong1,2, LIU Zhi-jie3, ZHENG Han3
(1.HunanUniversityofMedicine,Huaihua418000,China;2.The2ndXiangyaHospital,CentralSouthUniversity,Changsha410000,China;3.NationalInstituteforCommunicableDiseaseControlandPrevention,ChineseCenterforDiseaseControlandPrevention,Beijing102206,China)
Supported by grant from the Minister of Science and Technology, People’s Republic of China (Nos. 2013ZX10004221, 2013ZX10004216-001-002 and 81261120559)
Streptococcussuisis an important emerging zoonotic agent with high public health significance[1-2]. Humans can be infected by closing contact with pigs or pork products through skin wounds, or through consumption of raw pork[3-5]. In humans,S.suisusually produces meningitis and septic shock[6-7]. Recently, the number of humanS.suisinfections reported worldwide has increased significantly, with most cases originating in Asia[8-13].
S.suisis regularly identified using culture and biochemical assays, and a PCR test is used targetinggdhcoding for glutamate dehydrogenase[14-18]. However, evidence suggests that the primers used in thegdh-targeted PCR assay are not specific enough for identification ofS.suis[17], it failed to identify someS.suisisolates[18].
Loop-mediated isothermal amplification(LAMP), recently developed, is rapid, accurate, and cost-effective for detecting pathogens in clinical diagnostics[19-20]. LAMP is a rapid detection technique that does not require expensive thermocyclers as its results can be judged by naked eye[21]; it has become the preferred method for clinical and on-site diagnosis.
A LAMP assay targetingcps2Jgene for detectingS.suisserotype 2 strains is reported[22]; however, serotypes 1, 4, 5, 14, 16, and 24 in human cases cannot be detected using this assay[1,13,23-25]. Another LAMP assay targeting 16s rRNA genes for simultaneous detection ofStaphylococcusaureus,Streptococcuspneumoniae,S.suisandStreptococcusagalactiaewas reported[26]; however, the LAMP products need digestion with restriction endonuclease to identify the bacterial species. Here we chose another specific and conserved gene,dnaN, as the target gene; developed a rapid, sensitive, and highly specific LAMP assay to detectS.suis; and evaluated the assay’s performance.
Materials and methodsBacterial strains and DNA templates preparation
The genomic DNA of 607 bacterial strains were used in this assay (Table 1). Besides the 29 serotypes reference strains (not including the 4 controversial reference strains, serotype 20, 22, 26 and 33) of the 541S.suisstrains listed in Table 1, the rest 512 isolates were identified asS.suiswhen PCR test targeting bothgdhand 16S rDNA were positive[14,27], and confirmed by biochemical assay using API 20 STREP identification kit (bioMeá rieux, Marcy l'Etoile, France), as well as could be typed by Multilocus Sequence Typing (MLST) Scheme[28]. Genomic DNA fromS.suisstrain GZ1 was used to determine the LAMP sensitivity. The DNA was extracted using a Wizard genomic DNA purification kit (Promega, Madison, WI). The DNA templates for the other strains were prepared directly from bacterial colonies or bacterial broth culture using the boiling method. Briefly, bacteria grown overnight were harvested and suspended in 100 μL of distilled water and boiled at 100 ℃ for 10 min. The sample was immediately cooled on ice for 5 min and centrifuged at 13 000 r/min for 10 min. The supernatant, containing the DNA, was used as the template for PCR amplification and the LAMP assay.
Tab.1List of bacteria used in this study
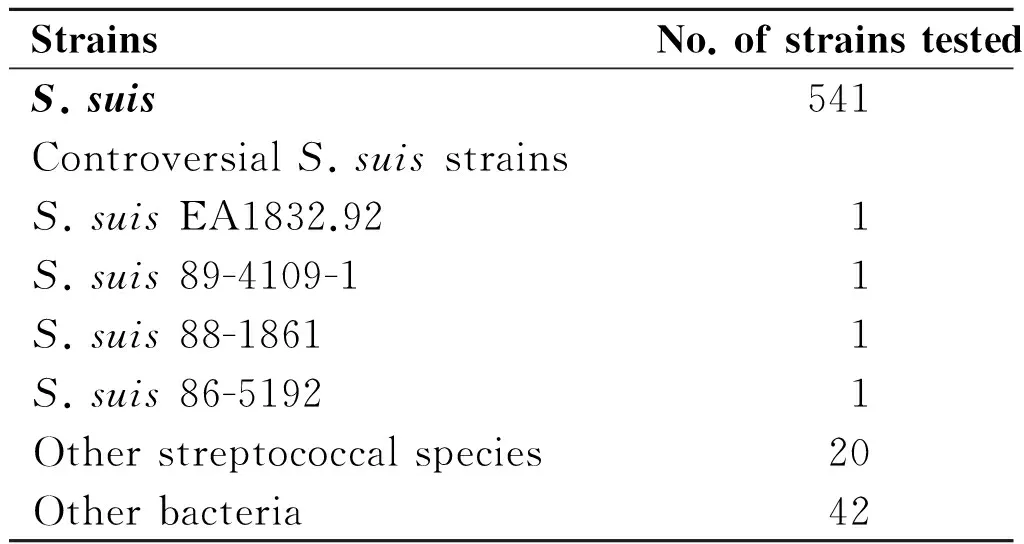
StrainsNo.ofstrainstestedS.suis541ControversialS.suisstrainsS.suisEA1832.921S.suis89-4109-11S.suis88-18611S.suis86-51921Otherstreptococcalspecies20Otherbacteria42
Specificity and conservation of thednaNgene inS.suis
We chose the genednaN, a core gene inS.suis, based on previous research in our laboratory[29]. ThednaN(SSGZ1_0002) andgdh(SSGZ1_ 0228) genes ofS.suisGZ1 (GenBank No. CP000837) were used and compared to other strains in the GenBank using the BLASTn program. Genomic DNA of all of theS.suisand non-S.suisstrains in Table 1 were tested using conventional PCR to determine the conservation and specificity ofdnaNinS.suis. Conventional PCR was performed with the outer primers (F3 and B3) from the LAMP assay targetingdnaN. The PCR cycling for the PCR reactions were: 94 ℃ for 5 min; followed by 30 cycles: 94 ℃ for 30 s, 63 ℃ for 30 s, and 72 ℃ for 30 s; and a final extension of 72 ℃ for 5 min using a thermocycler (Senso, Germany). Multiple sequence alignments ofS.suisand non-S.suisdnaNgene which available in GenBank were shown using DNAMAN version 5.2.2.
Primer design
A set of six primers targeting thednaNgene ofS.suiswere designed using PrimerExplorer V4 software (Eiken Chemical Co. Ltd., Tokyo, Japan) based on the conserved sequences determined by the alignment of thednaNgene sequences obtained from the GenBank. The primer set included two outer primers (F3 and B3) and two inner primers (FIP and BIP). To reduce the reaction time, we also designed LB and LF loop primers. The primers are shown in Table 2 and were synthesized by Sangon Biotech (Shanghai, China).
LAMP reaction
All LAMP reactions were performed with a Loopamp Kit (Eiken Chemical Co. Ltd., Tokyo, Japan) in a 25-μL mixture containing 1.6 μmol/L FIP and BIP primers (each), 0.8 μmol/L LF and LB primers (each), 0.2 μmol/L F3 and B3 primers (each), 20 mmol/L Tris-HCl (pH 8.8), 10 mmol/L KCl, 8 mmol/L MgSO4, 10 mmol/L (NH4)2SO4, 0.1% Tween 20, 0.8 mol/L betaine, 1.4 mmol/L deoxynucleoside triphosphates (dNTPs;each), and 1 μL Bst DNA polymerase (8 U/μL). The LAMP
Tab.2LAMP and conventional PCR primers used in this study to detect S. suis
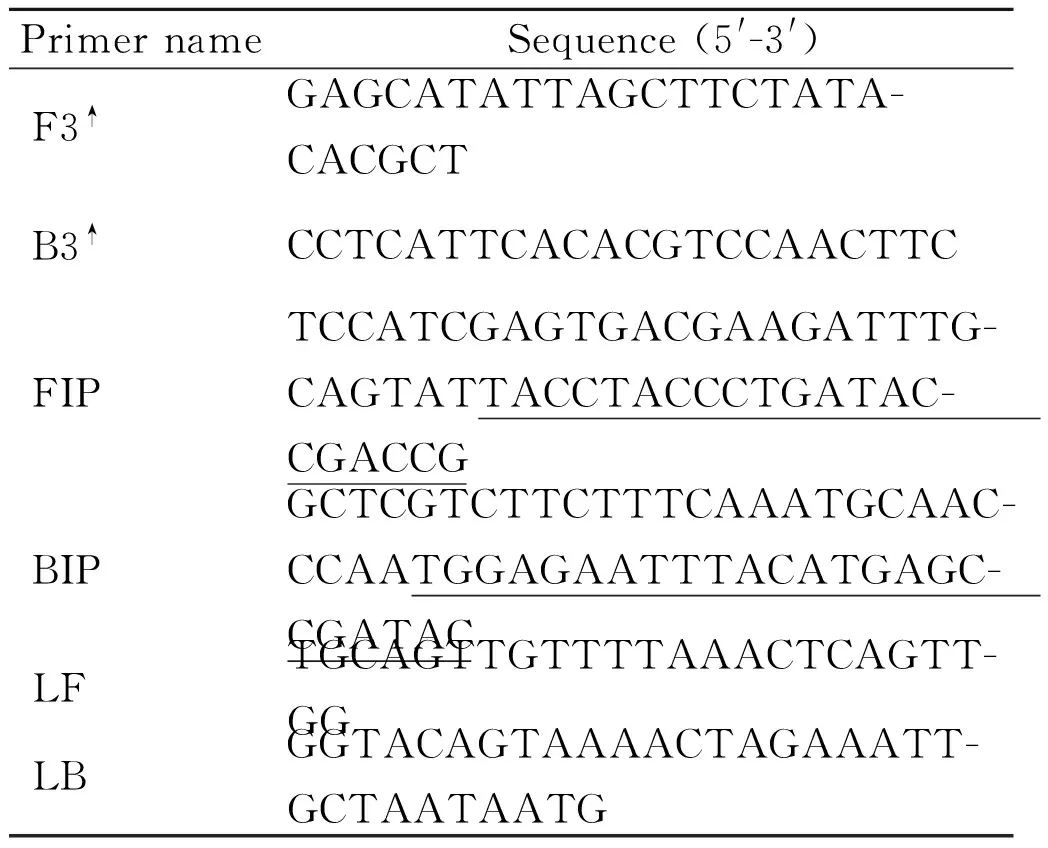
PrimernameSequence(5'-3')PositionF3↑GAGCATATTAGCTTCTATA-CACGCT706-730B3↑CCTCATTCACACGTCCAACTTC919-940FIPTCCATCGAGTGACGAAGATTTG-CAGTATTACCTACCCTGATAC-CGACCG800-827,741-761BIPGCTCGTCTTCTTTCAAATGCAAC-CCAATGGAGAATTTACATGAGC-CGATAC832-858,895-918LFTGCAGTTGTTTTAAACTCAGTT-GG769-792LBGGTACAGTAAAACTAGAAATT-GCTAATAATG862-892
Note:↑The primer pair F3/B3 was also used in the conventional PCR to confirm the specificity and conservation of the dnaN gene.
reaction was performed in different temperatures from 55 to 68 ℃ for 60 min using a LA-320C Loopamp real-time turbidimeter (Teramecs, Japan). The optimum conditions for the LAMP procedure were 63 ℃ for 60 min. Therefore, all of the mixtures were incubated at 63 ℃ for 60 min; followed by heating at 80 ℃ for 5 min to inactivate the reaction. A negative control (reaction mixture with distilled water instead of a DNA template) and a positive control (confirmed positive sample) were included.
Analysis of the LAMP products
The LAMP products were analyzed using three methods including a real-time turbidimeter, agarose gel analysis, and sight visualization. A LA-320C Loopamp realtime turbidimeter (Teramecs, Tokyo, Japan) was used to monitor the LAMP reaction based on the turbidity of magnesium pyrophosphate at 650 nm, a byproduct of the reaction. The turbidity threshold value for a positive sample was 0.1, and samples above this threshold value were considered as positive. After amplification, 5 μL of the LAMP product was further separated using 2% agarose gel electrophoresis stained with Goldenview and visualized under UV light. Using the calcein detection method, 1 μL of the calcein mixture (0.625 mmol/L calcein and 12.5 mmol/L MnCl2) was pre-added to the reaction mixture systems to create a total volume of 25 μL, and the color changes of the reaction mixtures were observed after amplification directly or visualized under UV light. When observed directly,the samples were considered positive if the solution turned green, judged as negative if the solution turned orange. The samples were considered positive if green fluorescence was observed and no fluorescence was judged negative using UV light.
Sensitivity and specificity of the LAMP assay
The sensitivity of the LAMP assay was tested usingS.suisGZ1. The detection sensitivity for genomic DNA and the DNA from the boiled cultured bacterial broth were determined. Genomic DNA was 2-fold serially diluted to concentrations of 1 pg, 500 fg, 250 fg, 125 fg, 62.5 fg, 31.25 fg, 15.625 fg, and 7.8125 fg/μL. One microliter of each dilution was used as the DNA template.
To assess the minimum number of cells detected by the LAMP assay using the boiling method,S.suisGZ1was grown to an optical density at 600 nm(OD600) of 0.6 in broth culture, 5-fold serial dilutions of the bacterial suspensions were performed. The number of colony forming units (CFU)/mL was determined by plating appropriate dilutions on Todd Hewitt Broth (THB) agar, incubating overnight at 37 ℃ in a 5% CO2atmosphere, and counting colonies. One milliliter serial dilution suspensions of bacteria were pelleted and suspended in 100 μL distilled water. DNA was released from the bacteria by boiling for 10 min followed by centrifugation at 13 000 r/min for 10 min. One microliter of supernatant was used as the DNA template. At least three independent experiments were performed to establish the sensitivity of the LAMP assay.
Genomic DNA of 62 non-S.suisstrains and four controversial reference strains in Table 1 were tested using LAMP to determine the specificity of the LAMP assay.
Evaluation of the LAMP assay
The evaluation of the LAMP assays was conducted in a water bath under the conditions described above. A total of 100S.suisstrains isolated from different parts of China including 29 serotype reference strains and 34 clinical strains isolated from patients or diseased pigs as well as 37 field strains isolated from healthy pigs recovered in different years were used in this study to evaluate the LAMP assay.
Results
Specificity and conservation of thednaNgene inS.suis
We compared the specificity and conservation of genes dnaN andgdh(regarded as the species-specific gene ofS.suis). Except for the four reference strains in serotypes 20, 22, 26, and 33, which were recently reported not to belong toS.suis[30], thednaNgene of GZ1 shared over 98.9% identities in otherS.suisstrains, while the lowest identity between GZ1 and otherS.suisstrains of thegdhgene was 94.2%. When compared to other bacterial species strains, the highest identity was 77% between thednaNgene of GZ1 andS.anginosusC238, and the highest identity was 82% between thegdhgene of GZ1 andS.pneumoniaeSPN994038. Through comparing of these two genes, thednaNgene is more conserved and specific than thegdhgene.
For each DNA sample of all of the 541S.suisstrains, a 235 bp amplicon was produced using conventional PCR targeting thednaNgene with F3 and B3 primer pairs. Since thednaNgene in the four reference strains of serotypes 20, 22, 26, and 33 shared less than 83% identity with the rest of otherS.suis, the conventional PCR reactions were negative for these four strains. All of the 62 non-S.suisstrains were negative using conventional PCR. Non-specific amplification bands were not observed in any of the samples tested. Therefore, we developed the LAMP assay forS.suistargeting thednaNgene.
Sensitivity and specificity of the LAMP assay
The detection limit of the LAMP assay for pure genomic DNA of GZ1 was 31.25 fg. The calculated copy number from the 2 038 034 bp full-length genome was 14 copies/μL. Given the boiling method is a more practical, easier, and faster way to acquire a DNA template, we also evaluated the sensitivity of this method. The detection limit of the LAMP assay for boiling method was 27 CFU/reaction. The sensitivity results of these two methods were analyzed using a real-time turbidimeter, agarose gel, and pre-added calcein shown in Figure 1.
Given the previous report shows the four reference strains of serotypes 20, 22, 26, and 33 do not belong toS.suis[30], as do the comparison results above; therefore we excluded these four strains from theS.suisstrains. To analyze the specificity of the LAMP assay, we aligned the primer region ofdnaN(between the primer F3 and B3) of strain GZ1, the four reference strains serotypes 20, 22, 26, and 33, and the top 10 non-S.suisstrains with the highest identity todnaNgene compared toS.suisGZ1. The alignments shown large divergence with the strain GZ1 in the primer region of the LAMP assay Figure 2. Simultaneously, high specificity was acquired when the LAMP assay was used with 62 non-S.suisstrains with no false positive amplifications observed. And as expected, the four reference strains considered not to beS.suis, serotypes 20, 22, 26, and 33, tested negative.
Evaluation of the LAMP assay
Our LAMP assay was used to detect 100S.suisstrains using a simple DNA preparation process, and the results were observed using direct visualization and with UV light. All of the strains tested positive by this LAMP assay.
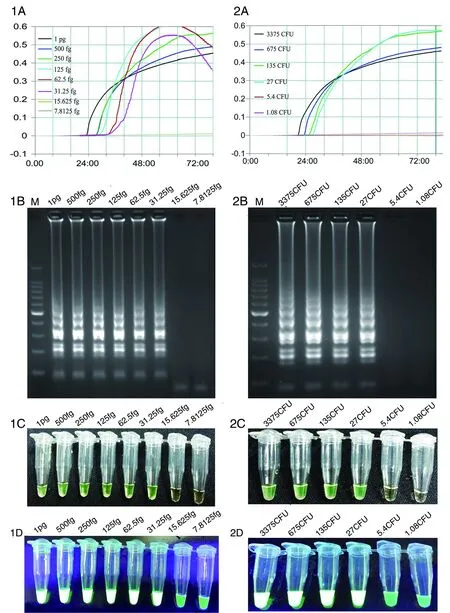
LAMP reactions using pure genomic DNA from strain GZ1 as the template detected by real-time turbidity (1A), electrophoresis of LAMP products (1B), and calcein visual observation using a color change visually (1C) and by a fluorescence assay under UV (1D). LAMP reactions detected using DNA prepared by the boiling method as the template of strain GZ1 by real-time turbidity (2A), electrophoresis of LAMP products (2B), and calcein visual observation for a color change (2C) and by a fluorescence assay under UV (2D). The limit of detection for pure S. suis genomic DNA was approximately 31.25 fg equal to 14 copies per reaction, and the limit of detection using the boiling method was 27 CFU per reaction. M indicates DNA marker in 1B & 2B.Fig.1 Sensitivity of dnaN-LAMP assay for the detection of S. suis
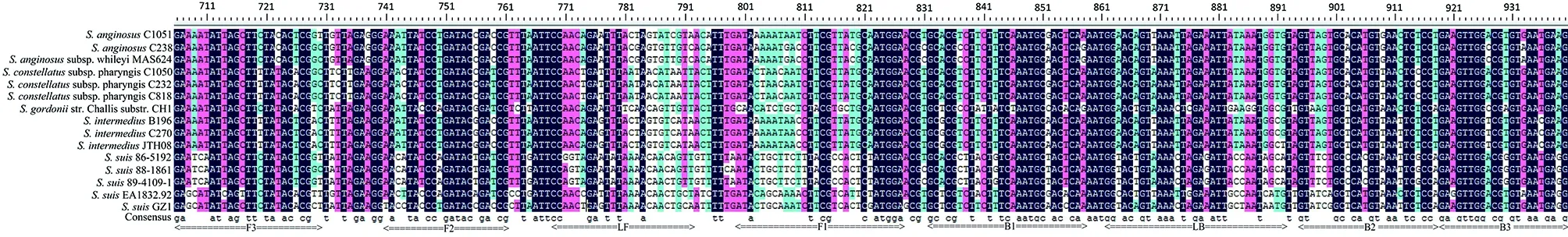
Multiple sequences of the primer region of dnaN (between the primer F3 and B3) of strain GZ1, four reference strains of serotypes 20, 22, 26, and 33, and the top 10 non-S. suis strains with the highest identity to the dnaN gene of S. suis GZ1 in the GenBank were aligned using DNAMAN version 5.2.2. Names and locations of the target sequences used as primers for the dnaN LAMP are shown at the bottom.Fig.2 Multiple sequence alignments of the primer region of dnaN gene
Discussion
S.suisinfection has become widespread in large-scale pig farms, causing huge economic losses and threatening public health due to its rapid spread and high mortality rates[2]. It is well known that the bacteriological methods available for the isolation and identification ofS.suisare tedious. PCR techniques and realtime PCR require a high-precision thermal cycler, and therefore, they are not adapted to diagnosingS.suisin basic clinical and field laboratories in rural areas. In contrast, the LAMP assay does not require sophisticated and expensive equipment, it just needs maintaining a constant temperature of 60 ℃-65 ℃ for 1 h is sufficient for the reaction[19]. These features demonstrate that the LAMP assay is suitable for the detection of S. suis in basic clinical and field laboratories in rural areas.
The choice of gene target was an important consideration in developing the LAMP assay. In our previous study[29], 85 isolates ofS.suiswere sequenced and compared, 876 genes were defined as the minimum core genome of the species. Among the 50 core genes with the lowest single-nucleotide polymorphisms (SNPs) density inS.suis, we chose the most conserved and specific gene dnaN encoding DNA polymerase III beta subunit forS.suisas the targeting gene. The bioinformatic comparison and the conventional PCR result showeddnaNis an ideal species-specific gene forS.suis.
In the sensitivity assay, the sensitivity of boiling method is a little lower than the method of pure genomic DNA, and may due to the toughS.suisGram-positive bacteria not releasing all of the genomic DNA using the boiling procedure. We tested 62 non-S.suisstrains and the four controversial reference strains to evaluate the specificity of thednaNLAMP assay for the bacteria, with the results showing that the specificity of the LAMP assay was 100%.
In conclusion, this study describes a rapid, simple, and reliable LAMP-based method that demonstrates high sensitivity and specificity for the identification ofS.suis. The assay is simple and does not require highly experienced technicians. And more importantly, the assay can be performed using a simple instrument such as a water bath at bedside or in rural areas or resource-limited laboratories. This assay has potential for use under field conditions for epidemic prevention, entry-exit inspection.
References
[1] Gottschalk M, Segura M, Xu J, et al.Streptococcussuisinfections in humans: the Chinese experience and the situation in North America[J]. Anim Health Res Rev, 2007, 8(1): 29-45.
[2] Gottschalk M, Xu J, Calzas C, et al.Streptococcussuis: a new emerging or an old neglected zoonotic pathogen?[J]. Future Microbiol, 2010, 5(3): 371-391. DOI: 10.2217/fmb.10.2
[3]Kay R, Cheng AF, Tse CY.Streptococcussuisinfection in Hong Kong[J]. QJM, 1995, 88(1): 39-47.
[4]Yu HJ, Liu XC, Wang SW, et al. Matched case-control study for risk factors of humanStreptococcussuisinfection in Sichuan Province, China[J]. Chin J Epidemiol, 2005, 26(9): 636-639.
[5]Takeuchi D, Kerdsin A, Pienpringam A, et al. Population-based study ofStreptococcussuisinfection in humans in Phayao Province in northern Thailand[J]. PLoS One, 2012, 7(2): e31265. DOI: 10.1371/journal.pone.0031265
[6]Arends JP, Zanen HC. Meningitis caused byStreptococcussuisin humans[J]. Rev Infect Dis, 1988, 10: 131-137.
[7]Huang YT, Teng LJ, Ho SW, et al.Streptococcussuisinfection[J]. J Microbiol Immunol Infect, 2005, 38(5): 306-313.
[8]Hui AC, Ng KC, Tong PY, et al. Bacterial meningitis in Hong Kong: 10-years' experience[J]. Clin Neurol Neurosurg, 2005, 107(5): 366-370.
[9]Ye C, Zhu X, Jing H, et al.Streptococcussuissequence type 7 outbreak, Sichuan, China[J]. Emerg Infect Dis, 2006, 12(8): 1203-1208.
[10] Yu H, Jing H, Chen Z, et al. HumanStreptococcussuisoutbreak, Sichuan, China[J]. Emerg Infect Dis, 2006, 12(6): 914-920.
[11]Ma E, Chung PH, So T, et al.Streptococcussuisinfection in Hong Kong: an emerging infectious disease?[J]. Epidemiol Infect, 2008, 136(12): 1691-1697. DOI: 10.1017/S0950268808000332
[12]Mai NT, Hoa NT, Nga TV, et al.Streptococcussuismeningitis in adults in Vietnam[J]. Clin Infect Dis, 2008, 46(5): 659-667. DOI: 10.1086/527385
[13]Kerdsin A, Dejsirilert S, Sawanpanyalert P, et al. Sepsis and spontaneous bacterial peritonitis in Thailand[J]. Lancet, 2011, 378(9794): 960. DOI: 10.1016/S0140-6736(11)60923-9
[14]Okwumabua O, O'connor M, Shull E. A polymerase chain reaction (PCR) assay specific forStreptococcussuisbased on the gene encoding the glutamate dehydrogenase[J]. FEMS Microbiol Lett, 2003, 218(1): 79-84.
[15]Silva LM, Baums CG, Rehm T, et al. Virulence-associated gene profiling ofStreptococcussuisisolates by PCR[J]. Vet Microbiol, 2006, 115(1-3): 117-127.
[16]Wei Z, Li R, Zhang A, et al. Characterization ofStreptococcussuisisolates from the diseased pigs in China between 2003 and 2007[J]. Vet Microbiol, 2009, 137(1/2): 196-201. DOI: 10.1016/j.vetmic.2008.12.015
[17]Tien le HT, Sugiyama N, Duangsonk K, et al. Phenotypic and PCR-based identification of bacterial strains isolated from patients with suspectedStreptococcussuisinfection in northern Thailand[J]. Jpn J Infect Dis, 2012, 65(2): 171-174.
[18]Gottschalk M, Lacouture S, Bonifait L, et al. Characterization ofStreptococcussuisisolates recovered between 2008 and 2011 from diseased pigs in Quebec, Canada[J]. Vet Microbiol, 2013, 162(2/4): 819-825. DOI: 10.1016/j.vetmic.2012.10.028
[19]Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA[J]. Nucleic Acids Res, 2000, 28(12): E63.
[20]Mori Y, Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases[J]. J Infect Chemother, 2009, 15(2): 62-69.
[21]Njiru ZK. Loop-mediated isothermal amplification technology: towards point of care diagnostics[J]. PLoS Negl Trop Dis, 2012, 6(6):692-694
[22]Zhang J, Zhu J, Ren H, et al. Rapid visual detection of highly pathogenicStreptococcussuisserotype 2 isolates by use of loop-mediated isothermal amplification[J]. J Clin Microbiol, 2013, 51(10): 3250-3256. DOI: 10.1128/JCM.01183-13
[23]Vilaichone RK, Vilaichone W, Nunthapisud P, et al.Streptococcussuisinfection in Thailand[J]. J Med Assoc Thai, 2002, 85 Suppl 1: S109-117.
[24]Nghia HD, Hoa NT, Linh le D, et al. Human case ofStreptococcussuisserotype 16 infection[J]. Emerg Infect Dis, 2008, 14(1): 155-157. DOI: 10.3201/eid1401.070534
[25]Kerdsin A, Oishi K, Sripakdee S, et al. Clonal dissemination of human isolates ofStreptococcussuisserotype 14 in Thailand[J]. J Med Microbiol, 2009, 58(Pt 11): 1508-1513. DOI: 10.1099/jmm.0.013656-0
[26]Huy NT, Hang le TT, Boamah D, et al. Development of a single-tube loop-mediated isothermal amplification assay for detection of four pathogens of bacterial meningitis[J]. FEMS Microbiol Lett, 2012, 337(1): 25-30. DOI: 10.1111/1574-6968.12002
[27]Marois C, Bougeard S, Gottschalk M, et al. Multiplex PCR assay for detection ofStreptococcussuisspecies and serotypes 2 and 1/2 in tonsils of live and dead pigs[J]. J Clin Microbiol, 2004, 42(7): 3169-3175.
[28]King SJ, Leigh JA, Heath PJ, et al. Development of a multilocus sequence typing scheme for the pig pathogenStreptococcussuis: identification of virulent clones and potential capsular serotype exchange[J]. J Clin Microbiol, 2002, 40(10): 3671-3680.
[29]Chen C, Zhang W, Zheng H, et al. Minimum core genome sequence typing of bacterial pathogens: a unified approach for clinical and public health microbiology[J]. J Clin Microbiol, 2013, 51(8): 2582-2591. DOI: 10.1128/JCM.00535-13
[30]Tien le, HT, Nishibori T, Nishitani Y, et al. Reappraisal of the taxonomy ofStreptococcussuisserotypes 20, 22, 26, and 33 based on DNA-DNA homology and sodA and recN phylogenies[J]. Vet Microbiol, 2013, 162(2/4): 842-849. DOI: 10.1016/j.vetmic.2012.11.001
Streptococcussuis(S.suis) is an important emerging zoonotic agent with high public health significance.S.suisis routinely identified using culture and biochemical assays and a PCR test targetinggdhcoding for glutamate dehydrogenase. Here we found thednaNgene to be more conserved and specific than thegdhgene inS.suis. Thus, thednaNgene was chosen as the target gene to design loop-mediated isothermal amplification (LAMP) assays for the rapid, specific, and sensitive detection ofS.suis. The LAMP procedure used 63 ℃ for 60 min. No false-positives were observed for the 62 non-S.suisstrains and the four controversial reference strains used to evaluate specificity of the assay. The limit of detection for pureS.suisgenomic DNA was approximately 31.25 fg, equal to 14 copies per reaction; the limit of detection using the boiling method was 27 colony forming units (CFU) per reaction. All of the 100S.suisstrains used in the evaluation assay were positive in the LAMP assay. This assay has potential for use in field conditions for epidemic prevention and entry-exit inspection.
dnaN; LAMP; rapid detection;S.suis
Zheng Han, Email: zhenghan@icdc.cn
2015-08-13;Revision accepted:2016-04-21
DOI:10.3969/j.issn.1002-2694.2016.06.007
