黄丹木姜子叶绿体基因组特征分析
2022-04-21刘潮韩利红彭悦樊瑞卿王宇默
刘潮 韩利红 彭悦 樊瑞卿 王宇默


摘要:【目的】分析黃丹木姜子( Litsea elongata)叶绿体基因组特征,为木姜子属物种鉴定、遗传多样性分析和资源保护提供理论参考。【方法】基于Illumina HiSeq 2000高通量测序平台对黄丹木姜子叶绿体基因组进行测序,利用GeSeq在线工具对叶绿体基因组进行注释,并利用REPuter、MISA、CodonW和IQ-TREE等生物信息学软件对其基因组结构、基因数目、序列重复、密码子使用偏性和系统发育进行分析。【结果】黄丹木姜子叶绿体基因组全长为154028 bp,具有典型的四分结构,编码126个基因,其中蛋白编码基因82个,rRNA基因 8个,tRNA基因 36个。叶绿体基因组的注释基因中,有9个基因含1个内含子,有3个基因含有2个内含子,其余基因均不含内含子;44个基因编码蛋白参与光合作用信号途径,21个基因编码蛋白构成了核糖体大小亚基。黄丹木姜子叶绿体基因组含有32对长序列重复和90个SSR位点,其中,正向重复和回文重复最多,均为12对,反向重复和互补重复分别为6和2对;95.56%的SSR位点位于单拷贝区[大单拷贝区(LSC)和小单拷贝区(SSC)],仅4.44%的SSR位点位于反向重复区(IR)。黄丹木姜子叶绿体蛋白编码基因GC含量为39.14%,GC3s为27.95%,平均有效密码子数(ENC)为49.04,说明其密码子偏性弱;相对同义密码子使用度(RSCU)大于1.00的密码子31个,其中13个以A结尾,16个以U(T)结尾。系统发育进化树分析结果显示,木姜子属的14个物种聚为两组,其中黄丹木姜子和10种木姜子属植物聚在一个组,与日本木姜子的亲缘关系最近。【结论】黄丹木姜子叶绿体基因组结构保守,偏好A或U(T)结尾的密码子,鉴定的SSR位点可用于物种鉴定和群体遗传学研究。
关键词: 黄丹木姜子;叶绿体基因组;SSR;密码子使用性;系统发育分析
中图分类号: S718.46 文献标志码: A 文章编号:2095-1191(2022)01-0012-09
Characteristics of chloroplast genome of Litsea elongata
(Wall. ex Nees) Benth. et Hook. f.
LIU Chao, HAN Li-hong*, PENG Yue, FAN Rui-qing, WANG Yu-mo
( College of Biological Resource and Food Engineering/Yunnan Engineering Research Center of Fruit Wine, Qujing Normal University, Yunnan, Qujing, Yunnan 655011, China)
Abstract:【Objective】 Research of Litsea elongata (Wall. ex Nees) Benth. et Hook. f. chloroplast genome possessed essential theoretical and practical significance for species identification, analysis of genetic diversity, and resource protection of Litsea. 【Method】The chloroplast genome of L. elongata was sequenced and annotated based on Illumina HiSeq 2000 high-throughput sequencing platform. The chloroplast genome was annotated using GeSeq. The genome structure,gene number, repeats, codon usage bias, phylogenetic development were analyzed by using the bioinformatics softwares such as REPuter, MISA, codonW and IQ-TREE. 【Result】The size of chloroplast genome of L. elongata was 154028 bp, with a typical quadripartite structure. The genome contained 126 genes, including 82 protein coding genes, 8 rRNA genes, and 36 tRNA genes.Among the annotated genes of chloroplast genome, nine genes contained one intron, three genes contained two introns. None of the remaining genes contained introns. Forty-four proteins were involved in the photosynthetic signal pathway, and 21 proteins constituted large/small subunit ribosome. The chloroplast genome of L. elongata contained 32 long repeats, and 90 simple sequence repeats (SSR). Among them, the forward repeats (12) and palindrome repeats (12) were the most, followed by the reverse repeats (6) and complementary repeats (2). 95.56% of the SSR loci were located in the single copy regions [large single copy region (LSC) and small single copy region(SSC)], and only 4.44% of SSR loci were located in the reverse repeat regions (IR). The guanine and cytosine (GC) content and synonymous third codons positions (GC3s) of chloroplast protein coding genes of L. elongata were 39.14% and 27.95%, respectively. And the average effective codon number (ENC) was 49.04, indicating that the codon bias of the chloroplast genome was weak. There were 31 codons with relative synonymous codon usage (RSCU) was greater than 1.00, of which 13 ended with A and 16 ended with U (T). Phylogenetic analysis showed that 14 Litseas pecies were clustered into two clades, and L. elongata gathered together with other ten Litsea species and shared the closest genetic relationship with L. japonica. 【Conclusion】The chloroplast genome structure of L. elongata is conservative and prefers codons ending in A or U(T). The identified SSR loci can be used for species identification and population genetics.06FB152F-66F4-4942-81C5-601FA2AEF053
Key words: Litsea elongata (Wall. ex Nees) Benth. et Hook. f.; chloroplast genome; SSR; codon usage; phylogenetic analysis
Foundation items: National Natural Science Foundation of China(32060710)
0 引言
【研究意义】黄丹木姜子[Litsea elongata (Wall. ex Nees) Benth. et Hook. f.]为樟科木姜子属(Litsea)常绿乔木,在我国华中、华东、华南和西南地区广泛分布,在尼泊尔、印度等国家也有分布,常生于山坡路旁或灌丛中,其木材和种子具有重要的应用价值。叶绿体是植物重要的细胞器,拥有相对独立的遗传系统,尤其是高等植物的叶绿体基因组具有较高的保守性,但不同物种间又存在局部的变异,故叶绿体基因组被广泛应用于植物分类和进化研究(Song et al.,2017a;Tian et al.,2019;Song et al.,2020;Zhang et al.,2021)。因此,开展黄丹木姜子叶绿体基因组特征分析,对木姜子属物种鉴定、遗传多样性分析及资源保护具有重要意义。【前人研究进展】叶绿体作为光合作用场所,在绿色植物生长发育和响应逆境过程中发挥作用(Pogson et al.,2015),因此,叶绿体基因组被广泛应用于樟科(Song et al.,2020)、双六道木属(Diabelia)(Wang et al.,2020)和木兰属(Magnolia)(Dong et al.,2021)等植物系统进化分析、物种鉴定、遗传多样性分析等方面。研究发现,大多数陆生植物的叶绿体基因组大小无明显差异(120~160 kb),并存在典型的四分结构,包括大单拷贝区(Large single copy ,LSC)、小单拷贝区(Small single copy,SSC)和2个反向重复区(Inverted repeat,IR)(Wicke et al.,2011)。目前,樟科(Song et al.,2020)、锦葵科(Wang et al.,2021)、杨属(Zong et al.,2019)、双六道木属(Wang et al.,2020)、木兰属(Dong et al.,2021)、辣椒属(刘潮等,2022)等多个科属的植物叶绿体基因组序列特征得到解析。大部分陆生植物进化过程中存在叶绿体基因组内的基因丢失、增加、重排和重复,叶绿体基因组中基因含量未发生显著变化(Wicke et al.,2011;Li et al.,2017;Song et al.,2017b;Li et al.,2021)。Song等(2020)利用120个樟科物种叶绿体基因组序列构建系统发育进化树,将樟科分为9个单系,木姜子属归为月桂—新木姜子组。基于解剖学、形态学和分子数据分析,发现木姜子属与山胡椒属植物在形态和分布上存在较多相似之处,基于核糖体ITS和叶绿体matK的分子系统分析显示,木姜子属与山胡椒属均表现为多系类群(Li and Christophel,2000;Li et al.,2004,2008)。【本研究切入点】目前,虽然樟科属间系统进化关系研究较多,而木姜子属内叶绿体基因组特征及系统进化关系有待进一步深入研究。【拟解决的关键问题】利用高通量测序技术对黄丹木姜子叶绿体基因组进行测序,以滇南木姜子(Litsea garrettii)叶绿体基因组为参照,对黄丹木姜子叶绿体基因组进行注释,并对基因组中序列重复、SSR位点及蛋白编码基因的密码子使用偏性进行分析,同时分析了木姜子属叶绿体基因组结构变异和系统发育关系,为黄丹木姜子及木姜子属植物资源开发与利用提供理论参考。
1 材料与方法
1. 1 试验材料
黄丹木姜子新鲜叶片采自浙江省温州市吹台山森林公园,样品置于硅胶中保存,存放于中国科学院西双版纳热带植物园(标本号XTBG-BRG-SY36963)。
1. 2 叶绿体基因组测序及注释
利用改良的CTAB法从叶组织中提取黄丹木姜子改成基因组DNA(李金璐等,2013)。基于Illumina Genome Analyzer HiSeq 2000测序平台完成叶绿体基因组测序。去除低质量测序片段,并使用GetOrganelle組装获得完整叶绿体基因组(Jin et al.,2020)。以滇南木姜子(L. garrettii)叶绿体体基因组(GenBank登录号MN698967)为参照,利用GeSeq(https://chlorobox.mpimp-golm.mpg.de/geseq.html)对黄丹木姜子叶绿体基因组进行注释。使用OGDRAW v. 1.3.1(https://chlorobox.mpimp-golm.mpg.de/OGDraw.html)绘制黄丹木姜子叶绿体基因组图谱(Greiner et al.,2019)。
1. 3 叶绿体基因组长序列重复和SSR位点分析
通过REPuter(Kurtz et al.,2001)分析长序列重复,搜索参数:最小重复长度为30 bp,序列同源性为90%,Hamming距离为3,同时分析了正向(Forward,F)、反向(Reverse,R)、互补(Complementary,C)和回文(Palindromic,P)重复。利用MISA-web在线工具(Beier et al.,2017)检测SSR,最小阈值为单核苷酸重复次数10,二核苷酸重复次数5,三核苷酸重复次数4,四核苷酸、五核苷酸和六核苷酸重复次数均为3。
1. 4 密码子偏性分析
使用CodonW 1.4.2(http://codonw.sourceforge.net/)和EMBOSS网站(http://emboss.toulouse.inra.fr/)的cusp软件分析黄丹木姜子叶绿体基因组中基因的有效密码子数(Effective number of codon,ENC)和相对同义密码子使用度(Relative synonymous codon usage,RSCU)(惠小涵等,2020)。06FB152F-66F4-4942-81C5-601FA2AEF053
1. 5 系统发育分析
从NCBI和LCGD数据库下载木姜子属13个物种的叶绿体基因组。将近缘类群樟(Cinnamomum camphora)和沉水樟(C. micranthum)设为外群物种(Song et al.,2020)。使用MAFFT(Katoh et al.,2019)进行多序列比对,通过IQ-TREE(Minh et al.,2020)使用最大似然法(Maximum likelihood,ML)构建系统发育进化树,建树模型为GTR+F+R2,步长值为1000。
2 结果与分析
2. 1 叶绿体基因组结构特征分析结果
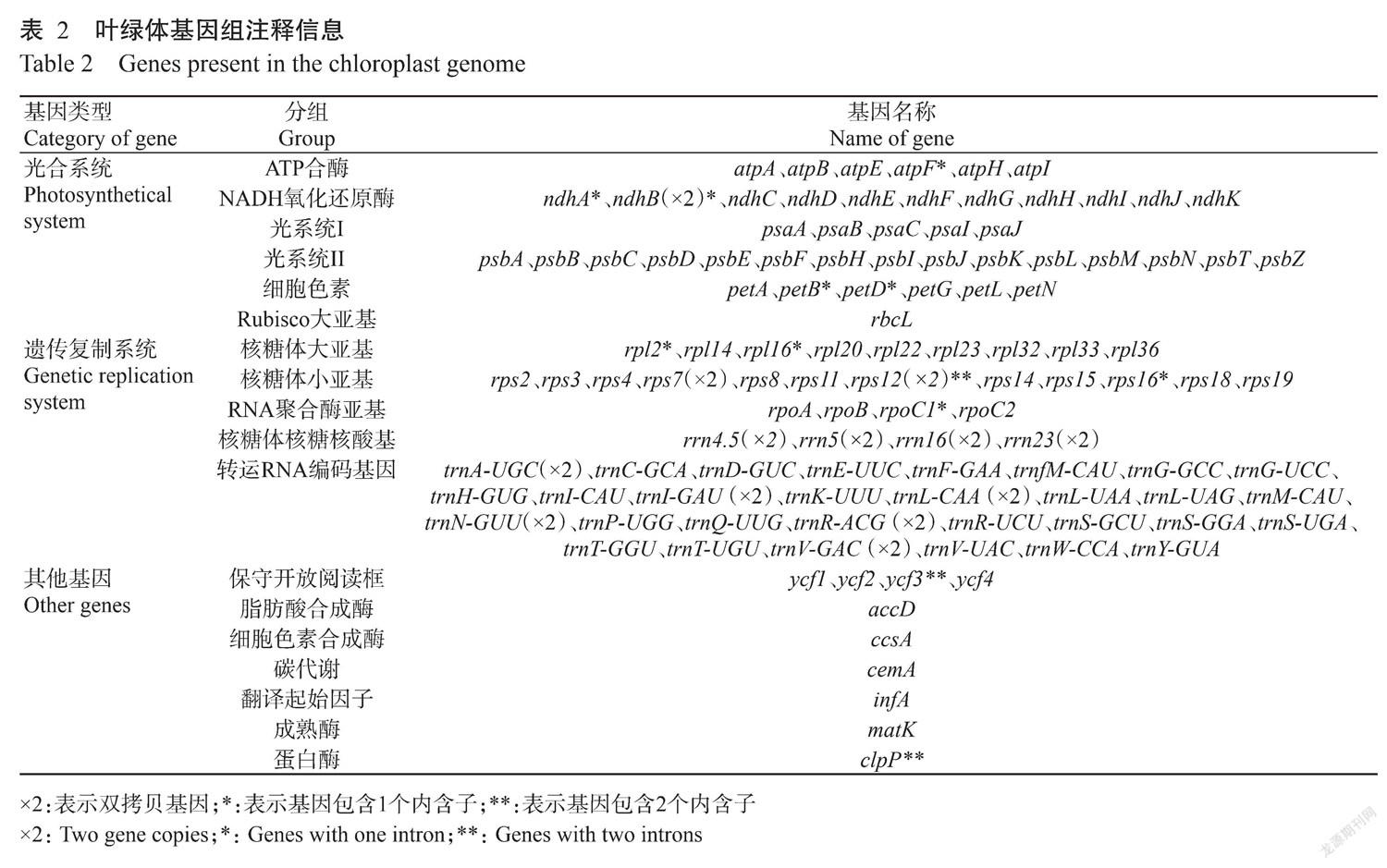
黄丹木姜子叶绿体基因组大小为154028 bp,具有典型的四分结构(图1),由LSC(93688 bp)、SSC(18852 bp)和2个IR(20744 bp)组成。黄丹木姜子叶绿体基因组共含126个基因,包含82个蛋白编码基因,8个rRNA基因和36个tRNA基因,其中13个基因为双拷贝基因(3个为蛋白编码基因,6个tRNA基因,4个rRNA基因)(表1);总GC含量为39.17%,四个分区中,IR的GC含量最高,其次为LSC,SSC最低;基因编码区及tRNA和rRNA编码区的GC含量均大于50.00%,蛋白编码基因的GC含量与叶绿体基因组接近。
黄丹木姜子叶绿体基因组注释基因中,有9个基因含1个内含子,有3个基因含2个内含子,其余基因均不含内含子。44个基因编码蛋白参与光合作用信号途径,21个基因编码蛋白构成了核糖体大小亚基(表2)。
2. 2 叶绿体基因组序列重复分析结果
黄丹木姜子叶绿体基因组序列含有32对(64条)长序列重复,其中正向重复和回文重复最多,均为12对(24条),反向重复和互补重复分别为6(12条)和2对(4条)(图2-A)。定位于LSC的长序列重复占32.81%,定位于IR的长序列重复占53.13%,定位于SSC的长序列重复占14.06%。最长的重复(48 bp)定位在LSC(图2-B)。长度为30 bp的重复数目最多,其中回文重复5对,正向和反向重复均为4对,互补重复为2对(图2-C)。
黄丹木姜子叶绿体基因组中有90个SSR位点,共12种类型(表3),其中,单核苷酸SSR位点数目最多,占72.22%,其次是二核苷酸SSR位点,占11.11%,三核苷酸重复4个,四核苷酸重复9个,五核苷酸和六核苷酸重复均為1个(表3)。二核苷酸SSR中AG/CT和AT/AT数目分别为4和6个,三核苷酸重复AAT/ATT数目为4个,四核苷酸重复AAAT/ATTT数目为5。77.78%的SSR位点位于LSC,17.78%的SSR位点位于SSC,仅4.44%的SSR位点位于IR。最长的SSR序列为单核苷酸和六核苷酸重复,大小均为18 bp。黄丹木姜子叶绿体基因组SSR序列偏好A/T碱基。69.66%的SSR位于基因间区,12.36%位于编码区序列上,17.98%位于内含子区(图3)。
2. 3 叶绿体基因组密码子偏性分析结果
黄丹木姜子叶绿体蛋白编码基因GC含量为39.14%,GC3s为27.95%,平均ENC为49.04,表明其密码子偏性较弱。蛋白编码基因中RSCU大于1.00的密码子为31个(图4和表4),其中13个以A结尾,16个以U(T)结尾,以G和C结尾的各有1个,表明黄丹木姜子叶绿体基因组蛋白编码基因更偏好A和U(T)结尾的密码子,与较低的基因组和蛋白编码基因GC含量一致。
2. 4 木姜子属植物系统发育分析结果
以樟和沉水樟为外类群,基于ML法利用叶绿体基因组序列构建系统发育进化树,结果显示,14个木姜子属植物聚为两大组(图5)。其中,潺槁木姜子、尖脉木姜子和木姜子以100%的支持率聚在组Ⅰ,黄丹木姜子和其他10种木姜子属植物聚在组Ⅱ,黄丹木姜子与日本木姜子的亲缘关系最近。
3 讨论
木姜子属植物包含大约400个种,分布于亚洲、中美洲、北美洲和太平洋岛屿的热带和亚热带森林中(Fijridiyanto and Murakami,2009)。黄丹木姜子分布广泛,其木材和种子均具有重要应用价值。植物叶绿体基因组的高保守性使其成为探究物种分类、遗传进化和谱系关系的理想工具(Song et al.,2017b)。随着高通量测序技术的发展,樟科植物(Song et al.,2020)、双六道木属植物(Wang et al.,2020)以及大花君子兰(Clivia miniata)(郑祎等,2020)、四川山胡椒(Lindera setchuenensis)(刘潮等,2021)、高良姜(Alpinia officinarum)(黄琼林,2021)等大量植物叶绿体基因组序列被解析。与木姜子属其他物种类似(Zhang et al.,2021),黄丹木姜子叶绿体基因组具有典型四分体结构,各区GC含量差异较大,其中IR的GC含量较高,这与rRNA的GC含量较高,且全部定位在这些区域密切相关(Xiao et al.,2020)。黄丹木姜子叶绿体蛋白编码基因偏好A或U(T)结尾的密码子,与樟(C. camphora)(秦政等,2018)、四川山胡椒(刘潮等,2021)和芝麻菜(Eruca sativa)(Zhu et al.,2021)等物种叶绿体基因组密码子使用性一致,可能与叶绿体基因组较低的GC含量有关,表明密码子偏好性可能受基因组GC含量的影响。密码子优化可提高基因翻译效率,密码子分析结果可为黄丹木姜子叶绿体转基因研究提供参考,根据叶绿体基因组的密码子偏好对目的基因进行优化,以提高基因转化和表达效率。
SSR常被用于物种鉴定、系统进化分析和群体遗传学研究(Li et al.,2021;Zhu et al.,2021)。长序列重复区域存在高度多态性,在植物基因表达和调控中起着重要作用。本研究从黄丹木姜子叶绿体基因组中鉴定到32对长序列重复和90个SSR位点,单核苷酸SSR占比较高,与鼎湖钓樟(Lindera chunii)(Tian et al.,2019)、四川山胡椒(刘潮等,2021)和天目木姜子(Litsea auriculata)(Zhang et al.,2021)等其他樟科植物重复序列特征相似,表明单核苷酸重复可能提供了更多的系统发育信息。黄丹木姜子叶绿体基因组中,53.13%的长序列重复分布在IR,95.56%的SSR位点分布在单拷贝区(LSC和SSC),69.66%的SSR位于基因间区,87.64%的SSR分布在基因的非编码区,与红景天属(Rhodiola)(Zhao et al.,2020)和木姜子属(Zhang et al.,2021)植物结果一致。此外,本研究还发现,基因组中含有较高比例的AG/CT、AT/AT、AAT/ATT和AAAT/ATTT,较高的A/T重复类SSR反映这些区域具有较高的遗传多样性,该结果与吊兰属(Chlorophytum)(Munyao et al.,2020)和山姜属(Alpinia)(Li et al.,2020)物种研究结果类似。本研究鉴定的重复序列可作为有价值的分子标记,用于黄丹木姜子的物种鉴定和群体遗传学研究。06FB152F-66F4-4942-81C5-601FA2AEF053
木姜子属植物在我国种类较多、分布广、使用价值高,具有广阔的开发利用前景,但由于相似种较多,木姜子属物种的识别和区分存在一定问题。基于形态学的分类方法存在一定的局限性(Li et al.,2000)。基于木姜子属叶绿体基因组序列构建的系统发育进化树显示,潺槁木姜子、尖脉木姜子和木姜子聚类在一组,其他木姜子属植物聚在另一组,与前人研究结果(Liu et al.,2020;Xiao et al.,2020;Zhang et al.,2021)一致,而与基于matK、ITS序列和表型构建的系统发育进化树不同(Li et al.,2004;Li et al.,2008)。尽管叶绿体基因组序列在物种分类和进化研究中显示出更高的分辨率和可靠性(Zhang et al.,2021),但对于相似种较多的木姜子属系统发育分析,需要更密集而全面的物种取样,同时结合基因组和转录组数据,才能得到更准确而系统的物种发育关系。叶绿体基因组特征及系统发育分析为黄丹木姜子物种鉴定、资源保护及开发利用等研究打下基础。
4 结论
黄丹木姜子叶绿体基因组结构保守,偏好A或U(T)结尾的密码子,鉴定的SSR位点可用于物种鉴定和群体遗传学研究。
参考文献:
黄琼林. 2021. 高良姜叶绿体基因组测序与特征分析[J]. 热带作物学报,42(1):1-6. [Huang Q L. 2021. Complete sequencing and analysis of chloroplast genome from Alpinia officinarum Hance[J]. Chinese Journal of Tropical Crops,42(1):1-6.] doi:10.3969/j.issn.1000-2561.2021.01.001.
惠小涵,程婷婷,柯卫东,郭宏波. 2020. 莲藕PPO基因密码子偏好性特征分析[J]. 江苏农业学报,36(2):438-446. [Hui X H,Cheng T T,Ke W D,Guo H B. 2020. Analysis on codon preference of PPO gene in lotus root[J]. Jiangsu Journal of Agricultural Sciences,36(2):438-446.] doi:10.3969/j.issn.1000-4440.2020.02.026
李金璐,王硕,于婧,王玲,周世良. 2013. 一种改良的植物DNA提取方法[J]. 植物学报,48(1):72-78. [Li J L,Wang S,Yu J,Wang L,Zhou S L. 2013. A modified CTAB protocol for plant DNA extraction[J]. Chinese Bulletin of Bo-tany,48(1):72-78.] doi:10.3724/SP.J.1259.2013.00072.
刘潮,唐利洲,韩利红. 2021. 四川山胡椒葉绿体基因组特征及山胡椒属系统发育[J]. 林业科学,57(12):167-174. [Liu C,Tang L Z,Han L H. 2021. Characterization of the chloroplast genome of Lindera setchuenensis and phylogenetics of the genus Lindera[J]. Scientia Silvae Sinicae,57(12):167-174.] doi:10.11707/j.1001-7488.20211217.
刘潮,韩利红,代小波,刘宸语. 2022. 辣椒属叶绿体基因组特征及进化[J]. 热带作物学报,43(3):447-454. [Liu C,Han L H,Dai X B,Liu C Y. 2022. Characteristics and phylogenetics of the complete chloroplast genomes of Capsicum Species[J]. Chinese Journal of Tropical Crops,43(3):447-454.] doi:10.3969/j.issn.1000-2561.2022.03. 002.
秦政,郑永杰,桂丽静,谢谷艾,伍艳芳. 2018. 樟树叶绿体基因组密码子偏好性分析[J]. 广西植物,38(10):1346-1355. [Qin Z,Zheng Y J,Gui L J,Xie G A,Wu Y F. 2018. Codon usage bias analysis of chloroplast genome of camphora tree(Cinnamomum camphora)[J]. Guihaia,38(10):1346-1355.]
郑祎,张卉,王钦美,高悦,张志宏,孙玉新. 2020. 大花君子兰叶绿体基因组及其特征[J]. 园艺学报,47(12):2439-2450.] [Zheng Y,Zhang H,Wang Q M,Gao Y,Zhang Z H,Sun Y X. 2020. Complete chloroplast genome sequence of Clivia miniata and its characteristics[J]. Acta Horticulturae Sinica,47(12):2439-2450.] doi:10.16420/j.issn.0513-353x.
Beier S,Thiel T,Münch T,Scholz U,Mascher M. 2017. MISA-web:A web server for microsatellite prediction[J]. Bioinformatics,33(16):2583-2585. doi:org/10.1093/bioinformatics/btx198.06FB152F-66F4-4942-81C5-601FA2AEF053
Dong S S,Wang Y L,Xia N H,Liu Y,Liu M,Lian L,Li N,Li L F,Lang X A,Gong Y Q,Chen L,Wu E,Zhang S Z. 2021. Plastid and nuclear phylogenomic incongruences and biogeographic implications of Magnolia s.l.(Magnoliaceae)[J]. Journal of Systematics and Evolution,161(8):107171. doi:10.1111/jse.12727.
Fijridiyanto I A,Murakami N. 2009. Molecular systematics of Malesian Litsea Lam. and putative related genera(Lauraceae)[J]. Acta Phytotaxonomica et Geobotanica,60(1):1-18. doi:org/10.18942/apg.KJ00005576218.
Greiner S,Lehwark P,Bock R. 2019. OrganellarGenomeDRAW(OGDRAW) version 1.3.1:Expanded toolkit for the graphical visualization of organellar genomes[J]. Nucleic Acids Research,47(W1):W59-W64. doi:10.1093/nar/gkz238.
Jin J J,Yu W B,Yang J B,Song Y,Depamphilis C W,Yi T S,Li D Z. 2020. GetOrganelle:A fast and versatile toolkit for accurate de novo assembly of organelle genomes[J]. Genome Biology,21(1):241. doi:10.1186/s13059-020-02154-5.
Katoh K,Rozewicki J,Yamada K D. 2019. MAFFT online service: Multiple sequence alignment,interactive sequence choice and visualization[J]. Briefings in Bioinformatics,20(4):1160-1166. doi:org/10.1093/bib/bbx108.
Kurtz S,Choudhuri J V,Ohlebusch E,Schleiermacher C,Stoye J,Giegerich R. 2001. REPuter:The manifold applications of repeat analysis on a genomic scale[J]. Nucleic Acids Research,29(22):4633-4642. doi:org/10.1093/nar/29.22.4633.
Li D M,Zhu G F,Xu Y C,Ye Y J,Liu J M. 2020. Complete chloroplast genomes of three medicinal Alpinia species: Genome organization,comparative analyses and phylogenetic relationships in family Zingiberaceae[J]. Plants (Basel),9(2). doi:10.3390/plants9020286.
Li J,Christophel D C,Conran J G,Li H W. 2004. Phylogenetic relationships within the ‘core Laureae(Litsea complex,Lauraceae) inferred from sequences of the chloroplast gene matK and nuclear ribosomal DNA ITS regions[J]. Plant Systematics and Evolution,246(1-2):19-34. doi:10.1007/s00606-003-0113-z.
Li J,Christophel D C. 2000. Systematic relationships within the Litsea complex(Lauraceae): A cladistic analysis on the basis of morphological and leaf cuticle data[J]. Australian Systematic Botany,13(1):1-13. doi:org/10.1071/SB98015.
Li J,Conran J G,Christophel D C,Li Z M,Li L,Li H W. 2008. Phylogenetic relationships of the Litsea complex and core Laureae(Lauraceae) using ITS and ETS sequences and morphology[J]. Annals of the Missouri Botanical Garden,95(4):580-599. doi:10.3417/2006125.9504.06FB152F-66F4-4942-81C5-601FA2AEF053
Li L,Hu Y,He M,Zhang B,Wu W,Cai P,Huo D,Hong Y. 2021. Comparative chloroplast genomes: Insights into the evolution of the chloroplast genome of Camellia sinensis and the phylogeny of Camellia[J]. BMC Genomics,22(1):138. doi:10.1186/s12864-021-07427-2.
Li Y,Zhou J G,Chen X L,Cui Y X,Xu Z C,Li Y H,Song J Y,Duan B Z,Yao H. 2017. Gene losses and partial deletion of small single-copy regions of the chloroplast genomes of two hemiparasitic Taxillus species[J]. Scientific Reports,7(1):12834. doi:10.1038/s41598-017-13401-4.
Liu C,Chen H H,Han L H,Tang L Z. 2020. The complete plastid genome of an evergreen tree Litsea elongata(Lauraceae: Laureae)[J]. Mitochondrial DNA Part B,5(3):2483-2484. doi:10.1080/23802359.2020.1778566.
Minh B Q,Schmidt H A,Chernomor O,Schrempf D,Woodhams M D,Von Haeseler A,Lanfear R. 2020. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era[J]. Molecular Biology and Evolution,37(5):1530-1534. doi:10.1093/molbev/msaa015.
Munyao J N,Dong X,Yang J X,Mbandi E M,Wanga V O,Oulo M A,Saina J K,Musili P M,Hu G W. 2020. Complete chloroplast genomes of Chlorophytum comosum and Chlorophytum gallabatense: Genome structures,comparative and phylogenetic analysis[J]. Plants(Basel),9(3):296. doi:10.3390/plants9030296.
Pogson B J,Ganguly D,Albrecht-Borth V. 2015. Insights into chloroplast biogenesis and development[J]. Biochimica et Biophysica Acta(BBA)-Bioenergetics,1847(9):1017-1024. doi:10.1016/j.bbabio.2015.02.003.
Song Y,Yao X,Tan Y,Gan Y,Yang J,Corlett R T. 2017a. Comparative analysis of complete chloroplast genome sequences of two subtropical trees,Phoebe sheareri and Phoebe omeiensis(Lauraceae)[J]. Tree Genetics and Genomes,13(6):120. doi:10.1007/s11295-017-1196-y.
Song Y,Yu W B,Tan Y,Liu B,Yao X,Jin J,Padmanaba M,Yang J B,Corlett R T. 2017b. Evolutionary comparisons of the chloroplast genome in Lauraceae and insights into loss events in the Magnoliids[J]. Genome Biology and Evolution,9(9):2354-2364. doi:10.1093/gbe/evx180.
Song Y,Yu W B,Tan Y H,Jin J J,Wang B,Yang J B,Liu B,Corlett R T. 2020. Plastid phylogenomics improve phylogenetic resolution in the Lauraceae[J]. Journal of Syste-matics and Evolution,58(4):423-439. doi:10.1111/jse. 12536.
Tian X,Ye J,Song Y. 2019. Plastome sequences help to improve the systematic position of trinerved Lindera species in the family Lauraceae[J]. PeerJ,7:e7662. doi:10.7717/peerj.7662.
Wang H X,Moore M J,Barrett R L,Landrein S,Sakaguchi S,Maki M,Wen J,Wang H F. 2020. Plastome phylogenomic insights into the Sino-Japanese biogeography of Diabelia(Caprifoliaceae)[J]. Journal of Systematics and Evolution,58(6):972-987. doi:10.1111/jse.12560.06FB152F-66F4-4942-81C5-601FA2AEF053
Wang J H,Moore M J,Wang H,Zhu Z X,Wang H F. 2021. Plastome evolution and phylogenetic relationships among Malvaceae subfamilies[J]. Gene,765:145103. doi:10.1016/ j.gene.2020.145103.
Wicke S,Schneeweiss G M,Depamphilis C W,Muller K F,Quandt D. 2011. The evolution of the plastid chromosome in land plants:Gene content,gene order,gene function[J]. Plant Molecular Biology,76(3-5):273-297. doi:10.1007/s11103-011-9762-4.
Xiao T,Xu Y,Jin L,Liu T J,Yan H F,Ge X J. 2020. Conflicting phylogenetic signals in plastomes of the tribe Laureae(Lauraceae)[J]. PeerJ,8:e10155. doi:10.7717/peerj.10155.
Zhang Y Y,Tian Y L,Tng D Y P,Zhou J B,Zhang Y T,Wang Z W,Li P F,Wang Z S. 2021. Comparative chloroplast genomics of Litsea Lam. (Lauraceae) and its phylogenetic implications[J]. Forests,12:744. doi:10.3390/f12060744.
Zhao D N,Ren Y,Zhang J Q. 2020. Conservation and innovation:Plastome evolution during rapid radiation of Rhodiola on the Qinghai-Tibetan Plateau[J]. Molecular Phylogenetics and Evolution,144:106713. doi:10.1016/j.ympev. 2019.106713.
Zhu B,Qian F,Hou Y,Yang W,Cai M,Wu X. 2021. Complete chloroplast genome features and phylogenetic analysis of Eruca sativa(Brassicaceae)[J]. PLoS One,16(3):e0248556. doi:10.1371/journal.pone.0248556.
Zong D,Gan P H,Zhou A P,Zhang Y,Zou X L,Duan A A,Song Y,He C Z. 2019. Plastome sequences help to resolve deep-level relationships of Populus in the family Salicaceae[J]. Frontiers in Plant Science,10:5. doi:10. 3389/fpls.2019.00005.
(責任编辑 陈 燕)06FB152F-66F4-4942-81C5-601FA2AEF053
