去势和非去势公猪背最长肌circRNA差异表达分析
2021-11-27邢宝松王璟陈俊峰马强任巧玲张家庆张华滑留帅孙加节曹海
邢宝松,王璟,陈俊峰,马强,任巧玲,张家庆,张华,滑留帅,孙加节,曹海
研究报告
去势和非去势公猪背最长肌circRNA差异表达分析
邢宝松1,王璟1,陈俊峰1,马强1,任巧玲1,张家庆1,张华1,滑留帅1,孙加节2,曹海3
1. 河南省农业科学院畜牧兽医研究所,河南省畜禽繁育与营养调控重点实验室,郑州 450002 2. 华南农业大学动物科学学院,广东省动物营养调控重点实验室/国家生猪种业工程技术中心,广州 510642 3. 河南兴锐农牧科技有限公司,信阳 465550
公猪去势可减少异味和打斗,但去势后产肉量和肌内脂肪沉积发生变化,其分子机制的解析对生产具有重要意义。近年来研究表明,环状RNA (circRNA)在肌肉发育中具有重要调控作用。为探究去势后circRNAs对背最长肌发育的调控,本研究选择6头淮南公猪,随机选择3头去势,当体重达130 kg左右屠宰,采集背最长肌样品,利用高通量测序筛选差异表达circRNAs (differentially expressed circRNAs, DECs)并进行KEGG功能富集分析。结合前期筛选的公猪去势相关miRNAs,构建DECs-miRNAs调控网络,最后使用猪骨骼肌卫星细胞验证候选circRNA表达谱及其与miRNA互作关系。结果表明,去势和非去势组背最长肌样品共获得5866个circRNAs,两组之间共有370个DECs (| log2Foldchange | > 1,p<0.8),KEGG富集分析表明,DECs来源母基因主要富集于肌肉发育、肌纤维类型转化、能量代谢等相关通路。构建的DECs-miRNA调控网络共包括69个circRNAs和8个miRNAs。选择circRNA_2241和circRNA_4237进行验证,结果发现这两个circRNAs真实存在且表达趋势与测序结果一致。进一步在猪骨骼肌卫星细胞初步验证circRNA_2241与miR-1互作关系,结果表明睾酮显著促进circRNA_2241表达,同时抑制miR-1表达。本研究结果提示circRNAs可能通过与miRNAs互作调控猪去势后背最长肌发育,从而为解析去势对肌肉发育调控的分子机制提供参考。
环状RNA;去势;公猪;背最长肌
在猪生产中,去势不仅减少公猪异味,还可减少打斗造成的经济损失。但去势后,公猪产肉量和肌内脂肪(intramuscular fat, IMF)含量与未去势公猪差异很大,去势公猪产肉量降低,脂肪沉积增加[1]。类似的,公牛去势后肉质性状例如IMF含量、大理石花纹、脂肪酸组成等都显著提高,但产肉量降低[2~4]。近年来,有研究比较了去势后背最长肌(longissimus dorsi, LD)和皮下脂肪mRNA、miRNA和lncRNA的表达变化。王璟等[5]比较了去势和非去势淮南公猪背最长肌转录组,共筛选到935个差异表达基因,KEGG富集到肌肉发育和脂质代谢相关通路。其中硬脂酰辅酶A去饱和酶1 (stearoyl-CoA desaturase-1, SCD-1)、激素敏感酯酶(hormone-sensitive lipase, HSL)、葡萄糖转运体4 (glucose transporter 4, GLUT4)等基因能同时或部分参与激素分泌、脂肪沉积和肌肉发育调控。Bai等[6]对比23周龄去势和非去势猪皮下脂肪组织miRNA表达谱,发现177个差异表达miRNAs,KEGG富集分析发现这些miRNAs参与肌细胞增殖、分化和凋亡和脂肪组织发育。Cai等[7,8]通过测序比较去势和非去势公猪皮下脂肪和背最长肌miRNAs表达差异,分别筛选到18个和7个差异表达miRNAs,其靶基因主要参与脂肪代谢和骨骼肌收缩。Wang等[9]在去势和非去势猪皮下脂肪组织筛选到18个差异表达lncRNAs,其靶基因与脂肪酸、胰岛素和脂肪细胞因子有关。Xing等[10]研究表明,去势和非去势猪背最长肌有385个差异表达lncRNAs,主要与雌激素受体的信号传导以及骨骼肌发育相关。虽然上述研究筛选了去势后背最长肌和皮下脂肪组织全转录组差异表达谱,但去势调控肌肉发育和脂质代谢的分子机制尚不清楚。
近期研究表明,环状RNA (circular RNA, circRNAs)参与肌肉发育和脂肪沉积调控,例如成肌细胞分化过程中circ-ZNF609表达量上调,可特异性抑制成肌细胞增殖[11]。线粒体分裂和凋亡相关circRNA (mitochondrial fission and apoptosis-related circRNA, MFACR)可通过抑制MTP18翻译减少心肌细胞死亡[12]。来源于鸡Supervillin基因的circSVIL通过竞争性吸附miR-203促进成肌细胞增殖和分化[13]。牛circFGF3可吸附miR-107,释放其对Wnt3a的抑制作用,进而促进成肌细胞分化[14]。circFUT10- miR-133a通路抑制成肌细胞增殖,并促进分化[15]。牛circHUWE1通过miR-29b-AKT3-AKT信号通路,促进成肌细胞增殖,抑制凋亡和分化[16]。circINSR通过海绵吸附miR-34a,减轻miR-34a对Bcl-2和CyclinE2的抑制,促进成肌细胞增殖减少细胞凋亡[17]。环状RNA SAMD4A通过miR-138-5p-EZH2促进前脂肪细胞分化[18]。CDR1as促进源自人脐带的间充质干细胞增殖和分化[19]。circFUT10通过let-7c-PPARGC1B促进牛脂肪细胞增殖抑制分化[20]。基于这些结果,为探究circRNAs在去势后猪肌肉发育中的调控机制,本研究利用高通量测序比较了去势公猪和未去势公猪背最长肌circRNA的表达差异,为进一步解析去势对肌肉发育调控机制提供参考。
1 材料与方法
1.1 实验动物
在河南兴锐农牧科技有限公司选择6头出生体重相近的半同胞淮南猪公猪,每2头来源于同一窝。于7日龄每窝随机选择1头去势,另1头相同部位进行伪手术处理,保证去势组和非去势组猪只所受手术应激一致。按照标准饲养流程饲喂,猪只体重达到130 kg左右(大约300~315日龄)屠宰,屠宰30 min内采集背最长肌样品(体右侧,第6~7肋骨),液氮保存。
1.2 RNA提取和测序建库
使用TRIzol (美国Invitrogen公司)分别提取6个背最长肌样品总RNA。利用琼脂糖凝胶电泳、Agilent 2100生物分析仪(美国安捷伦科技公司)和NanoDrop分光光度计(美国Nano-Drop科技公司)分析所提RNA的纯度、质量和完整性。RNA完整性数(RIN)大于8的样品用于构建文库。使用DNase I (美国QIAGEN公司)消化所提RNA,去除残留基因组DNA。使用Ribo-Zero™rRNA试剂盒(美国Epicentre公司)去除核糖体RNA。去势猪RNA样品和非去势猪RNA样品分别混合后测序。使用Illumina TruSeq™RNA样品制备试剂盒生成测序文库,在Illumina Hiseq 2500平台进行测序。
1.3 circRNA鉴定
原始数据(Raw data)去除接头和低质量数据得到有效数据(clean data),用TopHat2软件将有效数据与猪参考基因组(11.1)比对分析。通过find_circ软件鉴定circRNAs,其基本原理是:提取与基因组未比对上序列两端20 nt的anchor序列,反向拼接anchor序列获得短序列读段,将短序列读段再次与基因组进行比对,选取序列吻合且有GT-AG剪接位点的作为候选circRNA。将read count小于2的circRNA留作鉴定的circRNAs。与circBase数据库比对区分已知circRNAs和新发现circRNAs。进一步根据circRNAs在染色体的位置,分为反义,有义重叠,外显子,内含子和基因间五类。
1.4 circRNA表达分析
使用TPM(transcripts per kilobase of exon model per million mapped reads,每千个碱基的转录每百万映射读取的转录本数)对circRNAs进行归一化处理,计算每个circRNA在每个样品的表达量[21]。使用DESeq软件分析circRNAs在不同样品中的表达差异,差异表达circRNA (differently expressed circRNA,DEC)筛选条件为| log2Foldchange |≥1且p≤0.8[22]。利用Bowtie2软件鉴定circRNA的母源基因,对DECs母源基因进行GO和KEGG分析,<0.05视为有统计意义。
1.5 circRNA-miRNA网络图的构建
为进一步分析DECs的生物学功能,结合前期研究筛选的公猪背最长肌去势相关miRNAs[23],使用miRanda软件分析DECs与这些miRNAs之间的关系,保留种子区域没有错配,且能量< –18 kcal /摩尔的miRNAs。使用Cytoscape软件对DECs-miRNA互作网络进行绘图。
1.6 circRNA验证和定量分析
根据长度和表达量,选择circRNA_2241和circRNA_4237鉴定所筛选circRNA真实性和表达趋势。用RNase R (3 U/µg,美国Epicenter Biotechnologies公司)处理背最长肌提取的总RNA,1 µg RNA使用3 U的RNase R于37℃孵育15 min。根据circRNA_2241和circRNA_4237的剪切位点,设计特异性反向扩增剪切位点的引物,引物信息见表1,引物由上海生工生物工程有限公司设计合成。扩增后通过Sanger测序鉴定circRNA的真实性(上海生工生物工程有限公司)。
使用qRT-PCR检测circRNA_2241和circRNA_ 4237在去势和非去势猪背最长肌的表达水平,所用RNA与测序所用RNA相同。使用SYBR Green PCR试剂盒,扩增体系包括20 ng cDNA、10 µL 2×SYBR Premix ExTM和10 µmol/L上下游引物。qPCR扩增程序:95℃预变性5 min;95℃变性20 s,60℃复性20 s,72℃延伸20 s,40个循环。所有反应重复3次,并通过2–ΔΔCt法计算circRNAs相对表达量。
1.7 猪骨骼肌卫星细胞培养
circRNA_2241包含在DECs-miRNA互作网络中,所以选择circRNA_2241-miR-1做进一步验证。按文献[24]的方法分离猪骨骼肌卫星细胞。为验证去势对肌肉circRNA表达的影响,在猪骨骼肌卫星细胞中,通过添加不同浓度睾酮和不添加睾酮,分别模拟非去势组和去势组。当细胞达到70%~80%融合时,在培养基中添加不同浓度睾酮,对照组不添加睾酮,实验组睾酮添加量分别为10–9mol/L和10–10mol/L。添加睾酮48 h后收获细胞,检测睾酮对circRNA_2241和miR-1表达的影响。
1.8 统计分析
使用SPSS统计软件进行方差分析和显著性检验,所有数据均以mean±s.e.m.表示。<0.05表示差异显著,<0.01表示差异极显著。
2 结果与分析
2.1 circRNA特征分析
去势和非去势公猪背最长肌共获得5866个circRNAs,两组共有circRNAs为5205个(图1A)。这些circRNAs长度范围150 bp~99,406 bp,平均长度5494 bp (图1B)。这些circRNAs在全部染色体均有分布,6号染色体分布的circRNAs最多,占10.54%。X染色体和Y染色体分别有143个和9个,线粒体上仅有4个(图1C)。根据在基因组的位置,这些circRNAs中有义重叠最多(77%),其次是基因间区(14%),反义circRNAs和位于外显子区的circRNAs均为4%,内含子区最少,仅有1% (图1D)。
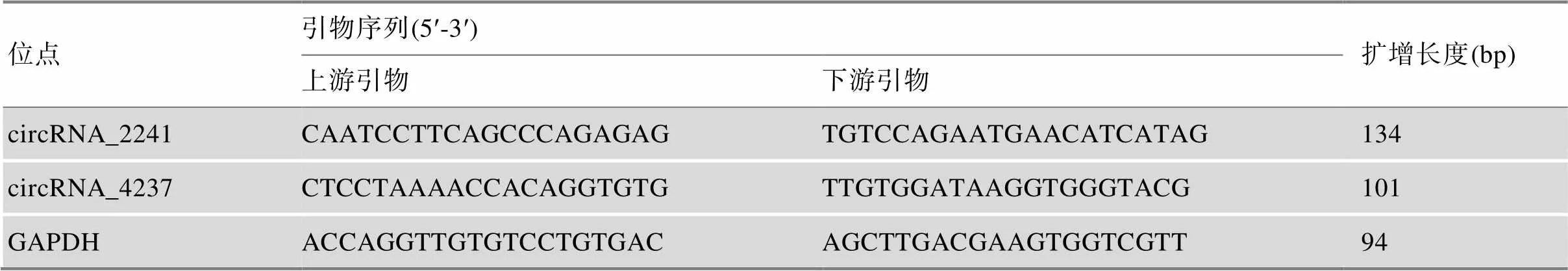
表1 circRNA引物序列信息

图1 去势和非去势组背最长肌鉴定circRNAs的特征
A:去势组和非去势组circRNAs差异;B:circRNAs分类;C:circRNAs长度;D:染色体分布情况。MT:线粒体。
2.2 circRNAs差异表达和功能分析
去势组和非去势组相比,共筛选到370个DECs,其中有217个上调,153个下调(| log2Foldchange | > 1,p< 0.8) (图2)。对这些DECs来源基因进行功能分析,GO分析主要富集的细胞组分为细胞器,生物学过程主要是各种代谢过程,分子功能主要富集于酶、蛋白质、核苷酸的结合(表2)。KEGG分析主要富集于肌肉发育、肌纤维类型转化和能量代谢相关通路,例如Wnt、泛素介导的蛋白水解、甲状腺激素、淀粉和蔗糖代谢、AMPK等信号通路(图3)。
2.3 circRNAs-miRNAs互作网络的构建和分析
为进一步解析这些DECs的功能,基于内源竞争RNA (competing endogenous RNAs,ceRNA)机制,结合前期获得的去势和非去势猪背最长肌差异表达的miRNAs,构建DECs-miRNA互作网络。如图4所示,网络图共富集69个circRNAs,8个miRNA,共234个edges,每个circRNAs最少有2个以上miRNA结合位点。其中miR-1靶circRNA最多,共38个,miR-133a-3p靶circRNA最少,共20个。其中circ_1060、circ_5230、circ_6457、circ_7356和circ_7733的miRNA结合位点最多,都有8个。

图2 去势和非去势组差异表达circRNAs火山图
图中一个点代表一个circRNA,每个点的横坐标值是该circRNA的log2Foldchange (Foldchange=去势组TPM/非去势组TPM),每个点的纵坐标为该circRNA在两组的–log10value。

图3 去势和非去势组DECs来源基因KEGG富集分析

表2 去势和非去势组DECs来源基因GO富集分析
互作网络图中富集到的69个circRNAs中,有9个位于基因间区,剩下60个circRNAs来源于42个编码基因。为了解这些circRNAs的功能,本研究对其来源mRNA进行了KEGG富集分析,结果发现这些mRNA显著富集于蛋白、脂质和糖类代谢通路(图5)。

图4 去势相关miRNA与DECs互作分析

图5 miRNA-DECs网络富集circRNAs的KEGG分析
2.4 circRNAs的验证
为验证所筛选circRNAs真实性,综合考虑转录本长度和表达量水平,本研究选择circRNA_2241和circRNA_4237进行验证,二者分别位于14号染色体和2号染色体。结果如图6所示,RNase R消化前后circRNA_2241和circRNA_4237扩增量略有差异,而线性的GAPDH在RNase R消化后没有扩增产物。进一步将扩增产物进行测序,证实这两个circRNA反向剪切位点真实存在(图7)。定量PCR分析表明,与非去势组相比,circRNA_2241在去势组表达量下调,circRNA_4237在去势组表达量上调,表达变化趋势和测序结果一致(图8)。

图6 RNase R消化法验证circRNA_2241和circRNA_ 4237
RNase R+:添加RNase R组,RNase R-:不添加RNase R组。
为验证预测的DECs-miRNAs互作关系,进一步选取circRNA_2241和miR-1进行验证。在猪骨骼肌卫星细胞添加不同剂量睾酮,qRT-PCR发现睾酮抑制miR-1表达,促进circRNA_2241表达(图10),睾酮对二者表达调控作用是相反的,提示circRNA_ 2241可能是miR-1靶基因。

图7 circRNA_2241和circRNA_4237剪切位点测序结果
红色箭头表示反向剪切位点。

图8 circRNA_2241和circRNA_4237在去势组和非去势组背最长肌的表达
*<0.05表示差异显著,**<0.01表示差异极显著。
3 讨论
以往的研究从mRNA、miRNA以及lncRNA等角度探讨了去势后肌肉发育变化的分子机制。近年来研究表明circRNAs也参与肌肉发育调控,所以本研究首次通过高通量测序比较去势和非去势淮南猪背最长肌circRNAs表达谱。测序共鉴定5866个circRNAs,主要为同义重叠类型,位于内含子区的最少,这与之前研究结果类似[25,26]。这些circRNAs大于2000 bp的最多,小于2000 bp中,200~400 bp最多。这些circRNAs分布于所有染色体,其中Y染色体和线粒体最少。

图9 睾酮对circRNA_2241和miR-1表达量的影响
**<0.01表示差异极显著。
肌肉生长受细胞数量和蛋白合成降解两个方面的调控,其中肌细胞数量在胚胎期已固定,与成肌细胞增殖分化相关,需要肌源性蛋白适时合成和降解。出生后肌肉肥大主要是蛋白质分解代谢和合成代谢动态平衡的过程[27]。之前研究表明,家畜去势后产肉量降低,即肌肉量减少。本研究中,KEGG富集分析结果提示,DECs可能通过泛素系统、甲状腺激素、Wnt、AMPK等信号通路参与去势后肌肉发育、肌纤维类型转化以及能量代谢的调控。
DECs来源基因最主要富集到的是泛素介导的蛋白水解信号通路,其中泛素蛋白酶体系统(ubiquitin- proteasomesystem, UPS)参与调控肌肉蛋白降解,泛素蛋白连接酶肌肉降解因子(muscle atrophy F-box, MAFbx)泛素化并降解分化蛋白,抑制肌肉蛋白合成[28]。肌肉环指蛋白1 (muscle RING finger 1, MuRF-1)通过泛素化导致集钙蛋白1 (calsequestrin 1, CASQ1)和肌球蛋白重链(myosin heavy chain, MYH)降解[29,30]。快慢肌是影响肉品质的重要因素[31],而MAFbx和MuRF-1降解快慢肌速度不同[32,33]。所以UPS系统一方面调控肌肉分化相关蛋白降解,调控肌肉分化,另一方面通过对快慢肌降解速度不同,间接调控肉品质。KEGG分析还富集到甲状腺激素信号通路,低水平甲状腺激素促进骨骼肌生长,高水平抑制骨骼肌生长[34]。甲状腺激素可促进糖和脂肪氧化,增加脂肪分解[35]。
Wnt信号通路中Wnt5a促进生肌性定向分化[36],Wnt10b抑制成肌细胞的成脂分化[37],Wnt5a促进慢肌纤维增多而Wnt11促进快肌纤维增加[38],myostatin通过Wnt/β-catenin信号通路调控慢肌纤维发育[39]。AMPK在肌肉能量代谢中起重要调控作用,AMPK活化促进GLUT4表达,促进其转运到细胞膜,提高酵解型肌纤维的葡萄糖吸收[40]。当肌肉中葡萄糖过量时,AMPK可降低磷酸化糖原合成酶(glycogen synthase, GS)活性抑制糖原合成[41]。此外,AMPK也参与调控骨骼肌的生长、肥大和再生[42]。
为进一步分析这些circRNAs的调控机制,基于ceRNA机制,结合前期获得的去势相关miRNAs,绘制了DECs-miRNA互作网络。该网络共富集69个DECs (占差异circRNAs的18.65%)和8个miRNAs,平均每个miRNAs靶向29个circRNAs。富集到的miR-1和miR-133都是肌肉特异性miRNAs,也是重要的非肌性基因表达的抑制因子。转录因子如成肌分化抗原(myogenic differentiation, MyoD)、肌细胞生成素(myogenin, MyoG)、血清应答因子(serum response factor, SRF)、肌肉增强因子2 (myocyte enhancer factor 2, AMEF2)都是miR-1和miR-133a的调节因子。miR-1靶基因有组蛋白脱乙酰基酶4 (histone deacetylase 4, HDAC4),转录因子YY1 (ying-yang 1)和调宁蛋白3 (calponin 3, CNN3),其中HDAC4是肌肉表达基因的转录抑制因子[43,44],YY1在肌肉基因转录中起负调控作用[45],CNN3调控肌动蛋白和肌球蛋白的重组和分解[46]。Hong等[47]发现猪miR-1的2个SNPs位点与I型和II型肌纤维面积和组成相关。miR-133靶向SRF促进成肌细胞增殖[43],调控骨骼肌分化过程中的主脑样蛋白1 (mastermind like transcriptional coactivator 1, MAML1)、胰岛素样生长因子1 (insulin like growth factor 1, IGF-1)和神经多嘧啶束结合蛋白(polypyrimidine tract binding protein 1, PTBP1)[48]。此外miR-133通过细胞外信号调节激酶(extracellular regulated protein kinases, ERK)促进成肌细胞分化[49]。谷氨酰胺乙酸(alpha glucosidase, GAA)通过miR-133a-3p和miR- 1a-3p激活AKT/mTOR/S6K信号通路,促进成肌细胞分化和骨骼肌生长[50]。Wnt/β-catenin信号通路通过诱导miR-133b和miR-206抑制Pax7表达,诱导肌源性分化[51]。由此可见,这69个DECs可能通过竞争性吸附miR-1、miR-133等参与肌细胞增殖、分化、肌纤维发育等过程,进而参与肌肉发育和肉质性状的调控。为进一步验证这69个DECs,对其来源基因进行KEGG分析,主要富集于碳水化合物、脂类和蛋白代谢相关通路,提示这些DECs参与去势后肌肉能量代谢的调控。
为验证高通量测序结果的准确性,根据circRNAs的长度和表达量,选择circRNA_2241和circRNA_ 4237进行验证。结果发现使用RNase R处理对circRNA_2241和circRNA_4237的表达量影响不显著,但RNase R处理后,GAPDH无扩增产物。同时使用反向引物扩增测序证实circRNA_2241和circRNA_4237确实以环状存在。RT-qPCR结果提示circRNA_2241和circRNA_4237在两组表达变化趋势和测序一致。同时在猪骨骼肌卫星细胞中,添加睾酮促进circRNA_2241表达,抑制miR-1表达。这与本研究中,非去势组circRNA_2241表达量高于去势组相一致。至于circRNA_2241是否能竞争性吸附miR-1还需要进一步构建载体,通过双荧光素酶系统进行验证。同时circRNA_2241通过吸附miR-1间接影响哪个靶基因参与肉质性状的调控,也需进一步的细胞试验验证。
综上所述,本研究通过高通量测序筛选了猪去势后背最长肌的DECs,构建了DECs-miRNAs互作网络,使用反向引物和RT-PCR证实筛选circRNAs的真实性,并通过细胞试验证实睾酮对circRNA_ 2241和miR-1的表达调控。这些结果提示circRNAs可能通过与miRNAs互作,参与去势后肌肉发育、肌纤维类型和能量代谢的调控,为解析去势后肌肉发育的分子调控机制提供了新思路。
[1] Trefan L, Doeschl-Wilson A, Rooke JA, Terlouw C, Bünger L. Meta-analysis of effects of gender in combination with carcass weight and breed on pork quality., 2013, 91(3): 1480–1492.
[2] Li Y, Wang MM, Li QF, Gao YX, Li Q, Li JG, Cao YF. Transcriptome profiling of longissimus lumborum in Holstein bulls and steers with different beef qualities., 2020, 15(6): e0235218.
[3] Zhou ZK, Gao X, Li JY, Chen JB, Xu SZ. Effect of castration on carcass quality and differential gene expression of longissimus muscle between steer and bull., 2011, 38(8):5307–5312.
[4] Zhang YY, Wang HB, Wang YN, Wang HC, Zhang S, Hong JY, Guo HF, Chen D, Yang Y, Zan LS. Transcriptome analysis of mRNA and microRNAs in intramuscular fat tissues of castrated and intact male Chinese Qinchuan cattle., 2017, 12(10): e0185961.
[5] Wang J, Hua LS, Chen JF, Zhang JQ, Ren QL, Bai HJ, Guo HX, Xu ZX, Xing BS, Bai XX, Cao H. Effect of castration on gene expression in Longissimus dorsi muscle of Huainan male pig by transcriptome analysis., 2019, 50(9): 1746–1758.
王璟, 滑留帅, 陈俊峰, 张家庆, 任巧玲, 白红杰, 郭红霞, 徐照学, 邢宝松, 白献晓, 曹海. 去势对淮南公猪背最长肌转录组的影响. 畜牧兽医学报, 2019, 50(9): 1746–1758.
[6] Bai Y, Huang JM, Liu G, Zhang JB, Wang JY, Liu CK, Fang MY. A comprehensive microRNA expression profile of the backfat tissue from castrated and intact full-sib pair male pigs., 2014, 15: 47.
[7] Cai ZW, Zhang LF, Chen ML, Jiang XL, Xu NY. Castration-induced changes in microRNA expression profiles in subcutaneous adipose tissue of male pigs., 2014, 55(2): 259–266.
[8] Cai ZW, Zhang LF, Jiang XL, Sheng YF, Xu NY. Differential miRNA expression profiles in the longissimus dorsi muscle between intact and castrated male pigs., 2015, 99: 99–104.
[9] Wang J, Hua LS, Chen JF, Zhang JQ, Bai XX, Gao BW, Li CJ, Shi ZH, Sheng WD, Gao Y, Xing BS. Identification and characterization of long non-coding RNAs in subcutaneous adipose tissue from castrated and intact full-sib pair Huainan male pigs., 2017, 18(1): 542.
[10] Xing BS, Bai XX, Guo HX, Chen JF, Hua LS, Zhang JQ, Ma Q, Ren QL, Wang HS, Wang J. Long non-coding RNA analysis of muscular responses to testosterone deficiency in Huainan male pigs., 2017, 88(9): 1451– 1456.
[11] Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I. Circ-ZNF609 is a circular rna that can be translated and functions in myogenesis., 2017, 66(1): 22–37.e29.
[12] Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao JN, Chen C, Yan KW, Ponnusamy M, Zhang YH, Li PF. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression., 2017, 24(6): 1111–1120.
[13] Ouyang HJ, Chen XL, Li WM, Li ZH, Nie QH, Zhang XQ. Circular RNA circSVIL promotes myoblast proliferation and differentiation by sponging miR-203 in chicken., 2018, 9: 172.
[14] Li H, Wei XF, Yang JM, Dong D, Hao D, Huang YZ, Lan XY, Plath M, Lei CZ, Ma Y, Lin FP, Bai YY, Chen H. circFGFR4 promotes differentiation of myoblasts via binding miR-107 to relieve its inhibition of Wnt3a., 2018, 11: 272–283.
[15] Li H, Yang JM, Wei XF, Song CC, Dong D, Huang YZ, Lan XY, Plath M, Lei CZ, Ma Y, Qi XL, Bai YY, Chen H. CircFUT10 reduces proliferation and facilitates differentiation of myoblasts by sponging miR-133a., 2018, 233(6): 4643–4651.
[16] Yue BL, Wang J, Ru WX, Wu JY, Cao XK, Yang HY, Huang YZ, Lan XY, Lei CZ, Huang BZ, Chen H. The circular RNA circHUWE1 sponges the miR-29b-AKT3 axis to regulate myoblast development., 2020, 19:1086–1097.
[17] Shen XM, Zhang XY, Ru WX, Huang YZ, Lan XY, Lei CZ, Chen H. circINSR promotes proliferation and reduces apoptosis of embryonic myoblasts by sponging miR-34a., 2020, 19: 986–999.
[18] Liu YJ, Liu HT, Li Y, Mao R, Yang HW, Zhang YC, Zhang Y, Guo PS, Zhan DF, Zhang TT. circular RNA SAMD4A controls adipogenesis in obesity through the miR-138-5p/ EZH2 axis., 2020, 10(10): 4705–4719.
[19] Yang LY, Bin Z, Hui S, Rong L, You BS, Wu PP, Han XY, Qian H, Xu WR. The role of CDR1as in proliferation and differentiation of human umbilical cord-derived mesenchymal stem cells., 2019, 2019: 2316834.
[20] Jiang R, Li H, Yang JM, Shen XM, Song CC, Yang ZX, Wang XG, Huang YZ, Lan XY, Lei CZ, Chen H. circRNA profiling reveals an abundant circFUT10 that promotes adipocyte proliferation and inhibits adipocyte differentiation via sponging let-7., 2020, 20: 491–501.
[21] Zhou L, Chen JH, Li ZZ, Li XX, Hu XD, Huang Y, Zhao XK, Liang CZ, Wang Y, Sun L, Shi M, Xu XH, Shen F, Chen MS, Han ZJ, Peng ZY, Zhai QN, Chen J, Zhang ZF, Yang RL, Ye JX, Guan ZC, Yang HM, Gui YT, Wang J, Cai ZM, Zhang XQ. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma., 2010, 5(12): e15224.
[22] Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2., 2014, 15(12): 550.
[23] Cai ZW, Zhang LF, Chen ML, Jiang XL, Xu NY. Castration-induced changes in microRNA expression profiles in subcutaneous adipose tissue of male pigs., 2014, 55(2): 259–266.
[24] Li JX, Su T, Zou C, Luo WZ, Shi GL, Chen L, Fang CC, Li CC. Long non-coding RNAregulates porcine satellite cell differentiation through/and., 2020, 8: 518724.
[25] Sun WX, Sun XC, Chu WW, Yu SG, Dong FL, Xu GF. circRNA expression profiles in human visceral preadipocytes and adipocytes., 2020, 21(2): 815–821.
[26] Xu TY, Wu J, Han P, Zhao ZM, Song XF. circular RNA expression profiles and features in human tissues: a study using RNA-seq data., 2017, 18(Suppl 6): 680.
[27] Mohammadabadi M, Bordbar F, Jensen J, Du M, Guo W. Key genes regulating skeletal muscle development and growth in farm animals., 2021, 11(3): 835.
[28] Chen K, Cheng HH, Zhou RJ. Molecular mechanisms and functions of autophagy and the ubiq-uitin-proteasome pathway., 2012, 34(1): 5–18.
陈科, 程汉华, 周荣家. 自噬与泛素化蛋白降解途径的分子机制及其功能. 遗传, 2012, 34(1):5–18.
[29] Gregorio CC, Perry CN, McElhinny AS. Functional properties of the titin/connectin-associated proteins, the muscle-specific RING finger proteins (MURFs), in striated muscle., 2005, 26(6–8): 389–400.
[30] Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I., 2004, 101(52): 18135–18140.
[31] Shen LY, Zhang SH, Wu ZH, Zheng MY, Li XW, Zhu L. The influence of satellite cells on meat quality and its differential regulation., 2013, 35(9): 1081–1086.
沈林園, 张顺华, 吴泽辉, 郑梦月, 李学伟, 朱砺. 骨骼肌卫星细胞对肉品质的影响及其分化调控. 遗传, 2013, 35(9): 1081–1086.
[32] Phillips SM, Glover EI, Rennie MJ. Alterations of protein turnover underlying disuse atrophy in human skeletal muscle., 2009, 107(3): 645–654.
[33] Salanova M, Schiffl G, Püttmann B, Schoser BG, Blottner D. Molecular biomarkers monitoring human skeletal muscle fibres and microvasculature following long-term bed rest with and without countermeasures., 2008, 212(3): 306–318.
[34] Millward DJ. Interactions between growth of muscle and stature: mechanisms involved and their nutritional sensitivity to dietary protein: the protein-stat revisited., 2021, 13(3): 729.
[35] Volke L, Krause K. Effect of thyroid hormones on adipose tissue flexibility., 2021, 10(1): 1–9.
[36] Reggio A, Rosina M, Palma A, Cerquone Perpetuini A, Petrilli LL, Gargioli C, Fuoco C, Micarelli E, Giuliani G, Cerretani M, Bresciani A, Sacco F, Castagnoli L, Cesareni G. Adipogenesis of skeletal muscle fibro/adipogenic progenitors is affected by the WNT5a/GSK3/β-catenin axis., 2020, 27(10): 2921–2941.
[37] Park YK, Park B, Lee S, Choi K, Moon Y, Park H. Hypoxia-inducible factor-2α-dependent hypoxic induction of Wnt10b expression in adipogenic cells., 2013, 288(36): 26311–26322.
[38] Anakwe K, Robson L, Hadley J, Buxton P, Church V, Allen S, Hartmann C, Harfe B, Nohno T, Brown AMC, Evans DJR, Francis-West P. Wnt signalling regulates myogenic differentiation in the developing avian wing., 2003, 130(15): 3503–3514.
[39] Jiang YL, Lian ZX, Li N, Wu CX. Myostatin: a negative regulator of skeletal muscle mass., 2000, 22(2): 119–121.
姜运良, 连正兴, 李宁, 吴常信. 肌肉生长抑制素基因的研究进展. 遗传, 2000, 22(2): 119–121.
[40] Kido K, Egawa T, Fujiyoshi H, Suzuki H, Kawanaka K, Hayashi T. AMPK is indispensable for overload-induced muscle glucose uptake and glycogenesis but dispensable for inducing hypertrophy in mice., 2021, 35(4): e21459.
[41] Liu XH, Bauman WA, Cardozo CP. Myostatin inhibits glucose uptake via suppression of insulin-dependent and -independent signaling pathways in myoblasts., 2018, 6(17): e13837.
[42] Thomson DM. The role of AMPK in the regulation of skeletal muscle size, hypertrophy, and regeneration., 2018, 19(10): 3125.
[43] Chen JF, Mandel EM, Thomson JM, Wu QL, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation., 2006, 38(2): 228–233.
[44] Backs J, Worst BC, Lehmann LH, Patrick DM, Jebessa Z, Kreusser MM, Sun Q, Chen L, Heft C, Katus HA, Olson EN. Selective repression of MEF2 activity by PKA- dependent proteolysis of HDAC4., 2011, 195(3): 403–415.
[45] Lu LN, Zhou L, Chen EZ, Sun K, Jiang PY, Wang LJ, Su XX, Sun H, Wang HT. A novel YY1-miR-1 regulatory circuit in skeletal myogenesis revealed by genome-wide prediction of YY1-miRNA network., 2012, 7(2): e27596.
[46] Tang ZL, Liang RY, Zhao SP, Wang RQ, Huang RH, Li K. CNN3 is regulated by microRNA-1 during muscle development in pigs., 2014, 10(4): 377–385.
[47] Hong JS, Noh SH, Lee JS, Kim JM, Hong KC, Lee YS. Effects of polymorphisms in the porcine microRNA miR-1 locus on muscle fiber type composition and miR-1 expression., 2012, 506(1): 211–216.
[48] Iqbal A, Ping J, Ali S, Zhen G, Juan L, Kang JZ, Ziyi P, Huixian L, Zhihui Z. Role of microRNAs in myogenesis and their effects on meat quality in pig - A review., 2020, 33(12): 1873–1884.
[49] Feng Y, Niu LL, Wei W, Zhang WY, Li XY, Cao JH, Zhao SH. A feedback circuit between miR-133 and the ERK1/2 pathway involving an exquisite mechanism for regulating myoblast proliferation and differentiation., 2013, 4(11): e934.
[50] Wang YJ, Ma JD, Qiu WL, Zhang JW, Feng SY, Zhou XK, Wang X, Jin L, Long K, Liu LY, Xiao WH, Tang QZ, Zhu L, Jiang YZ, Li XW, Li MZ. Guanidinoacetic acid regulates myogenic differentiation and muscle growth through miR-133a-3p and miR-1a-3p co-mediated Akt/ mTOR/S6K signaling pathway., 2018, 19(9): 2837.
[51] Cui S, Li L, Mubarokah SN, Meech R. Wnt/β-catenin signaling induces the myomiRs miR-133b and miR-206 to suppress Pax7 and induce the myogenic differentiation program., 2019, 120(8): 12740–12751.
Analysis of differentially expressed circRNAs in longissimus muscle between castrated and intact male pigs
Baosong Xing1, Jing Wang1, Junfeng Chen1, Qiang Ma1, Qiaoling Ren1, Jiaqing Zhang1, Hua Zhang1, Liushuai Hua1, Jiajie Sun2, Hai Cao3
Castration can reduce odor and fights in boars, but the carcass yield is reduced, and the intramuscular fat content is increased. Understanding its molecular mechanism is of great significance for production. Recent studies have shown that circular RNAs (circRNAs) play an important role(s) in the regulation of muscle development. To explore the effects of circRNAs on the development of longissimus dorsi (LD) muscle after castration, six Huainan male pigs were selected and three of which were randomly castrated. Six pigs were slaughtered when their body weight reached around 130 kg, and the LD muscle samples were collected. The differentially expressed circRNAs (DECs) were screened by high-throughput sequencing and functionally analyzed using the KEGG databases. DECs-miRNAs network was constructed, and the expression profiles of candidate circRNAs and their interactions with miRNAs were verified in porcine skeletal muscle satellite cells. The results showed that a total of 5866 circRNAs were obtained, and 370 DECs were identified in LD muscle between the castrated and intact groups (| log2Foldchange | > 1,p<0.8). KEGG enrichment indicated that the parental genes for the DECs were mainly enriched in the pathways associated with muscle development, muscle fiber type transformation, and energy metabolism. There were 8 miRNAs and 69 circRNAs enriched in the DECs-miRNA network. circRNA_2241 and circRNA_4237 were selected for verification, which showed that these two circRNAs really existed and their expression profiles were consistent with the sequencing results. Further, preliminary analysis showed that circRNA_2241 interacted with miR-1, and testosterone promoted circRNA_2241 but inhibited miR-1 expression. These results confirmed that circRNAs might participate in the regulation of LD muscle development after castration by interacting with miRNAs, thereby providing new materials and references for analyses on the molecular mechanisms of castration on the regulation of muscle development.
circRNAs; castration; male pigs; longissimus muscle
2021-04-27;
2021-07-28
国家自然科学基金青年基金项目(编号:31601927),河南省农业科学院科技创新创意项目(编号:2020CX18)和河南省重点研发与推广专项(编号:212102110010)资助[Supported by the National Natural Science Foundation of China (No. 31601927), Technological Innovation and Creative Project from the Henan Academy of Agricultural Sciences (No. 2020CX18) and Financial Budget Project of Henan Province (No. 212102110010)]
邢宝松,博士,副研究员,研究方向:猪的育种与管理。E-mail: bsxing@126.com
王璟,博士,副研究员,研究方向:遗传育种。E-mail: wangjing_0407@163.com
10.16288/j.yczz.21-162
2021/8/27 12:31:16
URI: https://kns.cnki.net/kcms/detail/11.1913.r.20210825.1518.002.html
(责任编委: 李明洲)
