植物ACCase基因的结构功能及表达调控研究
2021-02-07王保明颜士华谭晓风
王保明 颜士华 谭晓风
摘 要:乙酰辅酶A羧化酶(Acetyl-CoA carboxylase,ACCase)是催化脂肪酸合成的关键和限速步骤。该文介绍了ACCase的分类及结构特征,阐述了其在脂肪合成代谢中的作用和在除草剂中的应用,分析了它的表达调控及反馈机理,揭示了ACCase基因的克隆及表达鉴定,并展望了ACCase在植物育种中的应用前景。
关键词:植物;ACCase;基因;结构功能;表达调控
中图分类号 S364.3 文献标识码 A 文章编号 1007-7731(2021)01-0017-08
Study of the Structural Function and Expressing Regulation of ACCase Genes
WANG Baoming1,2,3 et al.
(1College of Agriculture & forestry Science and Technology, Hunan Applied Technology University, Changde 415000, China; 2College of Modern Agriculture, Linyi University of Science and Technology, Linyi 276000,China; 3“2011”Cooperative Innovation Center of Cultivation and Utilization for Non-Wood Forest Trees of Hunan Province (Central South University of Forestry and Technology),Changsha 410004, China)
Abstract: ACCase is the critical step and rate limiting step in fat synthesis. In this paper, its classification and structural features were introduced, and its roles in lipid metabolism and application in herbicides were elaborated. Moreover, its mechanism of expressing regulation and feedback were also analyzed, and the cloning and expression patterns of ACCase genes were revealed. Finally, its application and prospect in plant breeding were prospected.
Key words: Plant; ACCase; Gene; Structural function; Expressing regulation
1 乙酰輔酶A羧化酶的结构、分类和功能
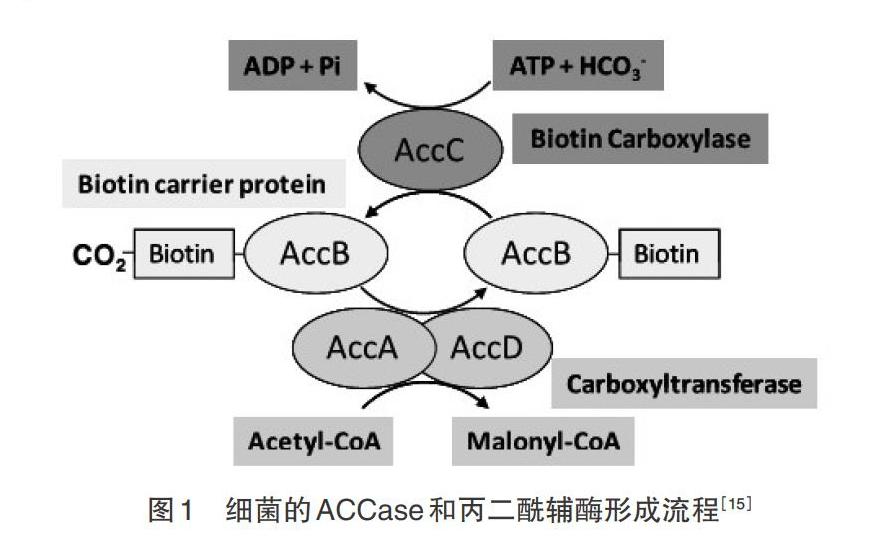
乙酰辅酶A(Acetyl coenzyme A,Acetyl-CoA)是碳进入油脂合成代谢途径的关键底物,而乙酰辅酶A羧化酶(Acetyl-CoA carboxylase,ACCase)能够催化Acetyl-CoA转化为丙二酰辅酶A(Malonyl CoA)而进入脂肪酸合成和油脂代谢途径。ACCase属于生物素Ⅰ酶,Malonyl CoA是脂肪酸合成和脂酰链延伸系统代谢的底物[1-2]。它在脂肪酸合成中作为C2单位供体,作为线粒体穿梭系统的调节因子,是脂肪酸氧化中一个代谢底物和蛋白活性的调控代谢物[3-5]。ACCase的催化反应不仅是关键步骤,也是限速步骤[4,6]。生物体内的ACCase包括异质型和同质型,其中,同质型ACCase含有生物素羧化酶(biotin carboxylase,BC)、生物素羧基载体蛋白(biotin carboxylase carrier protein,BCCP)和羧基转移酶(carboxyl transferase,CT)3个功能域[7]。异质型ACCase是植物从头合成脂肪酸的限速酶和关键酶,由BC、BCCP、α-CT和β-CT4个亚基组成[6,8-9]。其中,BC、BCCP和α-CT为核基因accC、accB以及accA编码,β-CT为质体基因accD编码。BC是糖异生、脂肪合成、氨基酸代谢和能量转换的关键酶。生物素共因子从碳酸盐捕获CO2,催化转运羧酸盐形成细胞代谢产物,它以共价键连接在BC亚基上,在ACCase羧化Acetyl-CoA的过程中作为羧基的中间载体,BC和CT各催化2个独立的半反应[9-17]:
第1个半反应:
E-biotin(BCCP)+[HCO-3]+Mg2+-ATP[]E-biotin-[CO-2]+Mg2+-ADP + Pi:biotin carboxylase;
第2个半反应:
E-biotin-CO2-(BCCP-CO2)+acetyl-CoA[]E-biotin(BCCP)+malonyl-CoA: carboxyltransferase。
BCCP作为活化羧基供体,在BC和CT之间摆动以传递羧基。其中,BC催化生物素羧化反应,含有ATP、Mg2+ 和CO2的结合点以及生物素辅基(共价键结合的生物素分子)和82个残基(包括Lys残基),在乙酰COA羧化反应中扮演中心角色。BCCP在BC催化下,CO2结合到生物素分子或羧基生物素;而它在CT的催化下,转移到乙酰CoA上成为Malonyl CoA。
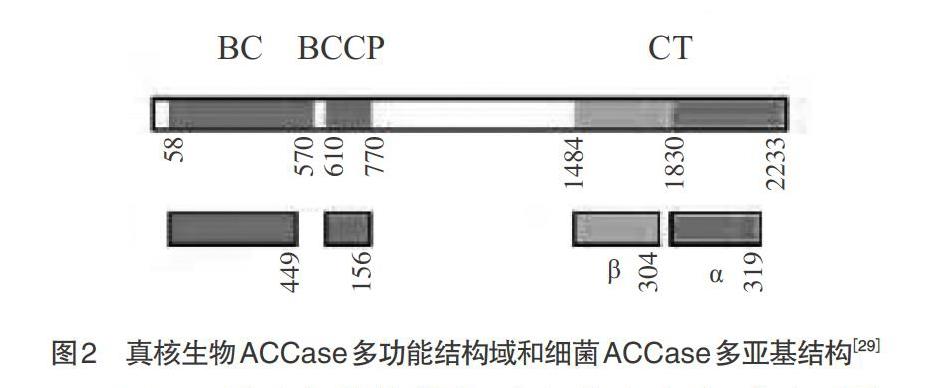
1.1 植物中ACCase的结构和分布 同质型ACCase即ACCase Ⅰ,也称多功能或真核型ACCase,分子量为220~280kDa,主要存在于酵母[18-19]、藻类[20]、动物[21-22]以及植物胞质溶胶中[23-25]。其基因序列与异质型ACCase的BC、BCCP、β-CT和α-CT组分相对应,形成多肽链上的3个功能域,活性状态下以同型二聚体形式出现,结构稳定难以解离。其结构形式为NH2-BC-BCCP-CT-COOH(见图2)[9]。1972年,研究者在菠菜叶绿体中发现了类似于大肠杆菌ACCase酶,直到1993年,才从豌豆的叶绿体中鉴定异质型ACCase[9,26]。异质型ACCase,又名原核类型ACCase,是BC、BCCP、α-CT和β-CT 4个亚基复合体。非活性条件下,这些亚基以单体形式存在;活性条件下,前2个亚基呈现同型二聚体,后2个是异型二聚体,两者以共价键相连构成CT催化域[27]。这类ACCase多存在于细菌、双子叶植物、禾本科单子叶植物的质体中[7,13]。异质型ACCase不稳定,易解离,它是由BC亚基同聚体与BCCP同聚体装配在一起,然后与α-CT和β-CT的异聚体松散连接[7,28]。其分子形式目前还不清楚,可能类似于细菌ACCase的(BCCP)4(BC)2(α-CT)2(β-CT)2 (见图2)[9,29]。
ACCase分布与植物器官、生长状态有关,小麦质体ACCase在中间部位较高,在根、叶片较低;而胞质溶胶ACCase在鞘科植物内含量较高[30]。在禾本科单子叶植物质体中,同质型ACCase与胞质溶胶中的不同。例如,小麦质体和胞质溶胶的同质型ACCase氨基酸序列同源性仅为67%,而其与玉米质体ACCase的同源性更高[31-32]。虽然玉米和小麦都是单子叶植物,但小麦胞质溶胶ACCase与双子叶植物胞质溶胶ACCase的同源性比与玉米质体ACCase的同源性高;油菜叶绿体中也存在同质型和异质型2种ACCase,即ACCase I 220kDa(存在于双子叶植物)和类似于大肠杆菌和拟南芥的多亚基异质型ACCase II复合体[33-34];其中ACCase II复合体中BCCP在氨基酸水平上与拟南芥有61%的一致性和79%的相似性[34]。
1.2 其他生物ACCase的結构、分布及特点 黏液球菌(Myxococcus xanthus)处于原核向真核生物演化期,其ACCase基因含有538氨基酸(aa)(ACCA)和573aa(ACCB)2个开放阅读框架(ORF),分别编码58.1kDa和61.5kDa2个BC亚基,前者与乙酰辅酶A羧化酶、丙酰辅酶A羧化酶以及丙酮酸羧化酶的BC亚基相似,含有ATP结合、固定CO2以及生物素结合模块。后者位于前者上游,与乙酰辅酶A羧化酶、丙酰辅酶A羧化酶、丙酮酸羧化酶的转羧酶和甲基丙二酰辅酶A脱羧酶高度相似,含有保守的羟基生物素结合位域和酰基辅酶结合域的CT亚基。它们形成1个两基因的操纵子[35]。在动物体内包括α和β 2种ACCase,其中,α-ACCase由ACACA基因编码,主要在肝脏、脂肪组织和乳腺中表达[4]。酵母、小鼠、鸡、人的α-ACCase的氨基酸序列约有90%相似性[36-37]。在转录过程中PⅠ、PⅡ启动子作用于α-ACCase基因[38]。β-ACCase由ACACB编码,在骨骼肌和心脏中表达,在肝脏和HepG2细胞中也有表达,有PⅠ、PⅡ2个启动子,它们的转录作用不同。人体中β-ACCase mRNA具有不同5′端的非翻译区[4]。人和大鼠的ACCase有2种同工酶,一个为265kDa,位于17号染色体上;另一个为275~280kDa,位于12号染色体的长臂上。在哺乳动物和人类中ACCase是一个多基因家族[37,39]。
1.3 ACCase的功能
1.3.1 在脂肪合成途径中的作用 脂肪酸合成起始于ACCase催化乙酰辅酶A产生Malonyl CoA,Malonyl CoA是脂肪酸合成的重要调控因素,它通过提高ACCase活性促进脂肪酸合成[40-41]。不同ACCase在脂肪酸合成中的作用不同。由异质型ACCase催化产生的Malonyl CoA用于脂肪酸从头生物合成,而由同质型ACCase催化产生的用于脂肪酸链延伸及类黄酮次生代谢产物合成。从头合成脂肪酸主要发生在质体,脂肪酸经过质膜在胞质溶胶和质体发挥作用[9]。动物体内α-ACCase在长链脂肪酸合成中催化限速反应并调节脂肪酸合成。在脂肪生成活跃组织中,Malonyl CoA的主要功能是作为长链脂肪酸从头合成碳链延长的C2供体从而合成三酰甘油和磷脂,并代替不同的脂肪乙酰辅酶A延长酶[42]。α-ACCase对胚胎早期发育具有重要作用[36]。β-ACCase主要调节脂肪酸氧化,其催化产物抑制脂肪酸氧化[43]。
1.3.2 反馈调节作用 拟南芥同质型ACCase基因在油菜质粒中过量表达,这种过量表达抑制了其他脂肪酸合成基因的表达而增加Malonyl CoA在前体中的积累[44]。14C标记的酰-酰载体蛋白加入外源脂肪酸后,在反馈抑制过程中酰-酰载体蛋白变化抑制Malonyl CoA的功能而导致脂肪酸合成降低。这暗示ACCase在脂肪酸合成的反馈调节中处于枢纽地位,是脂肪酸合成反馈调节的作用位点[45]。
1.3.3 在化学除草剂中的应用 ACCase是化学除草剂的重要靶标,其抑制剂是以ACCase为作用靶标的除草剂,能够抑制禾本科植物体内的脂肪酸合成。这类除草剂通过抑制真核型ACCase生成Malonyl CoA的羧化反应,进而抑制植物脂肪酸合成,有选择性地防除禾本科杂草[46-48]。例如,在菵草(Beckmannia syzigachne)中Trp1999Leu突变后对不同ACCase抑制除草剂的抗性产生差异,以此为依据选择合适的化学除草剂达到有效防治杂草的目的[49]。
2 ACCase的表达调控机理
由于异质型ACCase的BC、BCCP和α-CT在氨基端含有转移肽,它们的前体蛋白输送至叶绿体剪除转移肽后,组装成复合体才具有ACCase的催化活性[50]。植物器官中ACCase活性由复杂的机制调控,其基因表达调控主要包括转录调节、mRNA编辑和转录后调控[51]。
2.1 转录调节 转录调控主要通过发育诱导异质型ACCase的量与性质实现,细胞中脂肪酸合成达到活跃状态时或在之前,编码BC、BCCP、α-CT和β-CT的mRNA累积达到最大,随着脂肪酸合成减少,mRNA积累下降。mRNA最高积累发生在细胞快速分裂、生长和大量油脂积累中,与细胞生长有直接关系,并控制ACCase表达[51-52]。accA、accB、accC与质体中的accD互通信息并相互影响[52]。目前这3种核基因与质体基因的协调机制、增强子、转移因子还不清楚。质体在RNA转录至少包括质体编码聚合酶(Plastid-encoded polymerase,PEP)和核编码聚合酶(nucleus-encoded polymerase,NEP)2种聚合酶。PEP属于多亚基细菌型酶,含有大肠杆菌σ7启动子35(TTGaca)/–10(TAtaaT)元件;NEP含有T3/T7以及线粒体酶,多数NEP启动子由类似于植物线粒体的核心序列YRTA(type-Ia)控制,其中一些NEP启动子在YRTA模块上游含有GAA-box模块(type-Ib)。在质体中,光合基因具有PEP启动子(I类),非光合作用基因具有RNA聚合酶(II类),少数基因只由NEP转录(III类)[53-55]。PEP主要作用于叶绿体,在其周围含有大量的调控蛋白。如,在烟草质体基因组中,accD与psaI、ycf4、cemA、petA形成操纵子,在NEP型启动子控制下以多顺反子形式转录[56-59]。accD的表达决定因子是其5′UTR长度,5′UTR长度可能是NEP在质粒中转录效率的贡献因子[60]。
2.2 RNA编辑 叶绿体中RNA编辑对ACCD以及ACCase活性具有重要作用[61]。编辑后的酶具有活力,未编辑的则无活力。RNA编辑产生启始和终止密码,并改变编码序列。通常的变化是在三联体的第2个核苷酸位置上胞嘧啶(UCG)转变为尿嘧啶(UUG)[62]。一些植物的accD在相应位点没有亮氨酸密码子,而编辑后产生了亮氨酸密码子[61]。
2.3 转录后调节 ACCase亚基合成受转录后调控[53]。如,在烟草中以质体rRNA操纵子替代accD的操纵子,ACCase亚基的表达量和酶量增加[63],但是除了accD,其他基因的转录量均未增加。在野生型质体中,由于缺少accD亚基表达,其他3个亚基可能迅速降解,而在大量表达accD的植株中,这些过量表达能够装配进入ACCase[9]。ACCase表达量受基因转录控制,在装配中受蛋白质降解控制[8]。BC或BCCP同工酶2正向和反向表达都未改变ACCase亚基积累,这表明ACCase各亚基没有协调表达[9,53,64]。
3 ACCase的基因克隆及表达研究
3.1 ACCase的基因克隆
3.1.1 同质型ACCase基因的克隆 目前,已经从油菜[25]、苜蓿、小麦[32]、野生燕麦、黑麦草、拟南芥[65]和玉米中获得同质型ACCase基因的全长序列,它们长度均在10Kb以上,ORF为6700~7000bp,含有大量内含子。其中,油菜同质型ACCase至少由5个家族基因编码。硅藻基因组中含有2个ORF,较大的ORF长4.1Kb,位于较小ORF(2.2kb)的下游,中间含有73bp的内含子[66]。植物同质型ACCase的肽链上依次排列着BC、BCCP、α-CT功能域,它们由单一核基因编码。一些植物的种内外ACCase同源性存在差异。小麦质体ACCase的氨基酸序列与胞质ACCase的氨基酸序列同源性为67%,而它与玉米质体的高达80%[31]。植物ACCase最保守区域位于BC和BCCP中生物素羧化位点和生物素酰化位点周围。其中,生物素结合位点保守序列E(V/A)MK(M/L)为所有植物ACCase所共有[33]。在谷子中,2个编码2321个氨基酸ACCase cDNAs的亮氨酸/异亮氨酸位点可能是APPs和CHDs2类除草剂作用的关键位点[3]。1.8Kb长的BC功能域是ACCase最保守区域,也是生物素羧化位点、ATP结合位点[67]。
3.1.2 异质型ACCase亚基基因的克隆及结构分析 截至目前,已经克隆了大肠杆菌[12]、酵母菌[18]、豌豆[26]、小麦[30]、拟南芥[65]、烟草、马铃薯[68]、玉米[69]等生物,以及大豆[50]、油菜[33,70-71]、花生[72]、棉花[73]、蓖麻、棕榈[74]、麻疯树[75]等油料植物的异质型ACCase亚基基因。其中,大肠杆菌、拟南芥、大豆、花生和油菜[71]的已全部克隆,并且获得了BCCP和BC的晶状结构。大肠杆菌的BCCP和BC亚基基因共转录,其序列与老鼠丙酰辅酶羧化酶α-CT高度相似。生物素羧化酶活性和生物素(酰)化区域位于α-CT,而β-CT与老鼠丙酰辅酶羧化酶β-CT高度相似,并含有锌指模块CX2CX13-15CX2C催化羧基转移酶反应[8]。CT含有乙酰辅酶A结合域,其保守区域可能是CoA结合位点[10,13],在植物异质型ACCase中,叶绿体编码基因accD与大肠杆菌的同源,在禾本科植物中,accD基因或被截短,仅存一个短的C端区域,如水稻,或完全缺失,如小麦。其他核编码基因在胞质溶胶中转录翻译成前体蛋白,然后被转运到叶绿体中除去转移肽,与叶绿体中的β-CT加工组装成高分子量ACCase复合体[41,66]。
以探针筛选cDNA文库从酵母菌获得6个β-CT亚基,其中2个为全长cDNA[18]。拟南芥基因组异质型ACCase有2个基因编码的BCCP亚基,分别有1个基因编码BC、α-CT、β-CT亚基。马铃薯accD 5′末端包括典型的原核生物启动子类-35和-10序列TTGACA和TATCAA,ORF中包括乙酰辅酶A和羧基生物素结合位点、羧基转移酶催化位点[68]。在Brassica napus、Bassica rapa和Bassica oleracea中含有8个α-CT基因,这些基因含有9个外显子,其中,7个长度几乎相同,差异仅在第1个和最后1个外显子,最大差异出现在第2个内含子[70]。通过构建cDNA文库、EST测序、5′-RACE和3′-RACE分离出花生异质型ACCase accB1、accB2、accC、accA、accD和同质型ACCase基因。accD基因存在两处核苷酸编辑位点,初级结构高度保守[71]。油棕榈accD的氨基酸序列与其他植物的在N端差别大,含有CX2CX15CX2C锌指结构和(G/A)SMG(S/C)(V/A)VG、(V/L)(I/L)(I/M/L)V(C/S)(A/S)SGGARMQE、QM(A/G)KI(S/A)(S/A)(A/V)(L/S)、PT(T/A)GGVTAS(F/L)(G/A)(M/T)LGDIII(A/T)EP、FAGKR(V/I)IE(Q/E)(T/L)L5个保守模块[73]。
通过构建麻疯树cDNA文库和BAC文库,获得了α-CT、BCCP、BC、β-CT基因,它们的基因序列与其他植物的一致性较高。其中,α-CT、BCCP、BC是单拷贝核基因[75]。运用RT-PCR、RACE、基因组步移克隆出油菜ACCase的α-CT全长cDNA[73]。以简并引物获得序列为探针,从地中海拟无枝菌酸菌U32中获得的ORF为1797bp的accA基因[77]。从中棉35中克隆了5个编码异质型ACCase基因,其中,1个编码BC,1个编码BCCP,1个编码β-CT,2个编码α-CT,每个基因都是多拷贝[71]。从花生野生近缘种中克隆出accB1和accB2,它們的基因序列高度保守[78]。在陆地棉花中克隆出GhBCCP1、GhBC1、GhCTα2、GhCTβ,它们分别含有7、16、10、1个外显子[79]。在结构方面,异质型ACCase accA和accB的翻译产物在N端富含羟基化氨基酸Ser和Thr,疏水氨基酸Ala和Val,极性氨基酸Arg和Lys。这是转运肽的特点,在N端含有特征序列Met-Ala[33]。植物ACCase最保守区域是生物素羧化位点和生物素酰化位点周围,分别位于BC和BCCP,AMKLMN是保守的生物素位点,其中E(V/A)MK(M/L)是所有植物ACCase共有[33]。生物素(酰)化模块(C/G/M)-I-(V/I/L)-G-A-M-K-(M/L)-(M/E)-(N/I)在所有BCCP中高度保守[50]。如,拟南芥AtBCCP2和AtBCCP1的cDNA碳端生物素(酰)化模块(biotinylation)217EAMKLMNEIE226周围序列高度相似[9]。另外,BCCP中部区域的一个关键特征是高脯氨酸含量[50]。在accD 5端上游rbcL-accD基因间隔的核苷酸序列存在位点变化[80]。在桑科5属10个物种的rbcL-accD基因间隔发现了220处变化位点和16处插入或删除位点,但桑属rbcL-accD的序列却高度保守[81]。此外,利用数量位点(quantitative trait loci,QTL)研究发现:ACCase基因与控制脂肪酸和油脂QTL位点连锁影响玉米、燕麦种子中脂肪酸和油量变化[82-84]。在油桐中ACCase活性与种仁含油率具有正相关性[85]。
3.1.3 cDNA文库、转录组和基因组测序的应用 利用构建的油茶cDNA文库,克隆鉴定了油茶ACCase的accA、accB、accC和accD基因,分析它们的结构特点和表达模式[86-87]。利用油桐转录组克隆了ACCase亚基基因的cDNA序列[88]。利用基因组测序在Gossypium raimondii、G.arboreum、G.hirsutum、G.barbadense Gossypium中鉴定了4~8种ACCase BCCP基因的同源物[89]。基于雷蒙德氏棉、亚洲棉二倍体、陆地棉、海岛棉四倍体的基因组测序鉴定出棉花异质型ACCase核基因组编码的基因家族:24个BCCP基因、12个BC基因、11个α-CT基因,从而大大提高了ACCase基因的克隆效率[90]。
3.2 ACCase基因的表达及表达模式研究
3.2.1 表达研究 基因表达是在酶和调控序列的作用下基因转录成mRNA,经加工在核糖体协助下翻译出相应蛋白,在受体细胞经修饰而发挥特定生物学功能。表达产物主要包括RNA(tRNA、mRNA、rRNA、MicroRNA)、蛋白质、多肽等。基因表达产物可以揭示出基因表达生物学活性和生物学功能。通过构建共表达载体pHisAD,在大肠杆菌中表达豌豆质体CT,获得α-CT和β-CT多聚体的激活酶。该重组酶与来自豌豆叶绿体的大小相似,其催化活性与天然豌豆质体的相似[91]。将大豆异质型ACCase BC、BCCP、α-CT亚基基因cDNAs的转录产物输入到叶绿体中成为完整的ACCase,在大肠杆菌中产生特定抗体,利用抗体形成800kDa的BC/BCCP和600kDa的α-CT/β-CT复合体。将2个复合体一定比率混合,可以恢复ACCase活性,当把大豆(BC/BCCP)与豌豆(α-CT/β-CT)的复合体混合,可产生较高的ACCase活性[50]。ACCase BCCP和β-CT的mRNA和蛋白质表达分析暗示发育叶片和种子是脂肪酸高效合成器官[33]。Arabidopsis中的AtBCCP1和AtBCCP2异构体,其中,前者在叶、根、花、长角果表达,而后者主要在花、种子等生殖器官中大量表达。花和长角果中的mRNA积累反映了在授粉和贮存积累油脂的需要。前者可能为看家基因,出现在所有器官,后者主要表现在生殖器官[28]。马铃薯accD在叶片、茎、根、块茎的转录表达,表明它是一个看家基因[68]。异质型ACCase是所有组织中的关键蛋白。但是在花后40d种子中的转录比在花、真叶、纤维中转录水平要低,可能是因为这个时期脂肪酸合成速度降低的缘故[72-73]。
3.2.3 表达模式研究 基因表达具有时间特异性和空间特异性,前者多与细胞或个体的特定分化、发育阶段相适应,为阶段特异性;后者由细胞在组织器官的分布差异决定,为细胞特异性或组织特异性。在花生中,异质型ACCase和同质型ACCse基因在所有组织表达,它们的RNA表达差别很大。种子发育中accC、accA、accD的mRNA在花后60d大量表达,其中accC和accA表达模式相似,在叶片和种子大量表达,而accD在叶片中的表达比其他组织中多;同质型ACCase在茎和花中表达较多。accB2在花后50~70d大量表达,种子中表达量最多,而accB1在花后80d以前表达量变化较大,在叶片中表达多[73]。半定量和实时定量PCR发现油棕榈accD和accC协调表达,它的表达对异质型ACCase水平、种子油产量至关重要[74]。在麻疯树叶片中accD的表达比其他3个基因约高6倍,这可能是叶片富含叶绿体的缘故。在受粉后42d胚中的表达最大,暗示油脂合成达到高峰[75]。番茄果实颜色由绿色、变色、浅红、成熟红变化,转录和翻译中accD基因转录丰度增加,成熟达到最大,暗示在果实成熟中大量需求油脂,并贮存基质满足果实成熟大量合成类胡萝卜素的需求[92]。陆地棉花的中GhBCCP1、GhBC1、GhCTα2、GhCTβ在各组织中表达,与油脂积累具有正相关性并增加油含量[79]。
4 ACCase在植物育种中的应用及前景
植物油脂合成主要受质体ACCase控制。通过农杆菌转化系统过量表达同质型ACCase和叶绿体转化系统过量表达异质型ACCase,实现了数量调控脂肪酸的合成。在细菌(Escherichia coil)中通过提高乙酰辅酶A羧化酶活性促进脂肪酸合成[41]。通过构建细菌多顺反子,使脂肪酸含量大幅增加[41,93]。在大肠杆菌中过量表达ACCase,不但大幅地提高了丙二酰辅酶A的表达量,而且显示出协同效应,增加乙酰辅酶A的利用性,有助于脂肪酸合成[94]。
4.1 同质型ACCase的应用 同质型ACCase主要位于胞质溶胶,是一个大于200kDa的二聚体,催化产生的丙二酰CoA用于超长脂肪酸延伸。将油菜种子贮藏蛋白特异表达启动子与拟南芥同质型ACCase基因ACC1融合,在大豆Rubisco SSU转移肽作用下,定向将胞质溶胶ACCase导入于油菜叶绿体,大幅度提高了成熟种子的ACCase活性,并增加了种子含油量并改变了脂肪酸组成。将同质ACCase定位质体,既保护免于细胞质蛋白质代谢影响,也不受控制质体活性的调节抑制,这是转基因植物产油率高的原因[44,95]。将ACCase定位于淀粉体并过量表达,不但增加脂肪酸合成,而且大幅增加了三酰基甘油含量[95]。相反,反义表达抑制油菜同质型ACCase活性显著降低成熟种子的含油量[96]。可见,同质型ACCase的遗传操作是一个合理的应用策略。
4.2 异质型ACCase的应用 质体中NEP和PEP参与转录,通过同源重组转化烟草质体,在烟草中以NEP和PEP启动子替代accD的启动子,能够提高脂肪酸含量,延长叶片寿命,提高种子产量[56]。这项技术已经成功地应用到油料植物中[63,98]。反之,减少ACCase亚基因的表达会影响植物的生长和发育。同源重组剔除烟草部分accD基因及上游DNA序列,会导致烟草植株叶片出现白绿色斑点或叶片数目减少[99]。利用特异启动子过量表达ACCase亚基基因能够增加种子油含量[79]。然而,在烟草中组成型启动子正向和反向表达烟草的BC亚基,BC的表达变化没有影响BCCP表达。又如,分别构建组成启动子与BCCP2反义表达、napin种子启动子与BCCP2基因正义表达载体,在反义表达拟南芥植株中的轉录量表达增加,但没有明显的表型变化;在正义表达的拟南芥发育种子中虽然BCCP2表达量增加,但是子代种子的脂肪酸含量反而比野生拟南芥种子要低很多。这暗示BC和BCCP亚基不是ACCase积累的限制因子[100]。总之,由于这些调控、抑制、反馈作用使得ACCase的转基因应用是一个十分复杂的过程。
參考文献
[1]Konishi T,Shinohara K,Yamada K,et al.Acetyl-coA carboxylase in higher plants:most plants other than Gramineae have both the prokaryotic and the eukaryotic forms of this enzyme[J].Plant Cell Physiol.,1996,37:117-122.
[2]任波,李毅.大豆种子脂肪酸合成代谢研究进展[J].分子植物育种,2005,3(3):301-306.
[3]赵虎基,王国英.植物乙酰辅酶A羧化酶的分子生物学与基因工程[J].中国生物工程杂志,2003,23(2):12-16.
[4]韩春春,王继文,魏守海.乙酰辅酶A羧化酶(ACC)的结构和功能[J].安徽农业科学,2006,34:413-414,416.
[5]卢善发.植物脂肪酸的生物合成与基因工程[J].植物学通报,2000,17(6):481-491.
[6]Thelen J J and Ohlrogge J B.Metabolic engineering of fatty acid biosynthesis in plants[J].Metabolic Engineering,2002,4:12-21.
[7]Kondo H,Shiratsuch K,and Yoshimoto T,et al.Acetyl-CoA carboxylase from Escherichia coil:gene organization and nucleotide sequence of the biotin carboxylase subunit[J].Proc.Natl.Acad.Sci.USA,1991,88:9730-9733.
[8]Ohlrogge J,Browse J.Lipid Biosynthesis[J].The plant cell,1995,7:957-970.
[9]Sasaki Y,Nagano Y.Plant acety-CoA carboxylase:structure,biosynthesis,regulation,and gene manipulation for plant breeding[J].Biosci.Biotechnol.Biochem.,2004,68(6):1175-1184.
[10]Chapman-Smith A and Cronan J E.Symposium:Nutrition,biochemisty and molecular biology of biotin molecular biology of biotin attachment to proteins[J].J.Nutr.,1999,129:477S–484S.
[11]Alban C,Jullien J,Job D,et al.,Isolation and characterization of biotin carboxylase from Pea Chloroplasts[J].Plant Physiol.,1995,109:927-935.
[12]Li Shyr-Jiann,Cronan J E Jr.The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-coA carboxylase[J].The Journal of Biological Chemistry,1992,267(2):855-863.
[13]Li S J,Cronan J E.The gene encoding the biotin carboxylase subunits of pea acetyl-CoA carboxylase[J].J.Biol.Chem.,1992,267:16841-16847.
[14]Acetyl-CoA carboxylase.Wikipedia,http://en.wikipedia.org/wiki/Acetyl-CoA carboxylase.
[15]Alves J,Westling L,Peters EC,et al.Cloning,expression,and enzymatic activity of Acinetobacter baumannii and Klebsiella pneumoniae acetyl-coenzyme A carboxylases[J].Anal.Biochem.,2011,417(1):103-111.
[16]Wan Minxi,Liu Peng,Xia Jinlan,et al.The effect of mixotrophy on microalgal growth,lipid content,and expression levels of three pathway genes in Chlorella sorokiniana[J].Applied Microbiology and Biotechnology,2011,91(3):835-844.
[17]Nikolau B J,Ohlrogge J B,Wurtele E S.Plant biotin-containing carboxylase[J].Arch.Biochem.Biophys.,2003,414:211-222.
[18]Hasslacher M,Tvessa AS,Paltauf F,et al.Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipids metabolism[J].J.Biol.Chem.,1993,268:10946-10952.
[19]Walid A F,Chirala S S,Wakil S J.Cloning of the yeast FAS3 gene and primary structure of yeast Acetyl-CoA carboxylase [J].Proc.Natl.Acde.Sci.USA,1992,89:4534-4538.
[20]Roessler P,Ohlrogge J B.Cloning and characterization of the gene that encodes acetyl-coenzyme A carboxylase in the Alga Cyclotella cryptica[J].J.Biol.Chem.,1993,268:19254-19259.
[21]Lopez-Casillas F,Bai D H,Luo X N,et al.Structure of the coding sequence of acetyl-coenayme A carboxylase[J].Proc.Natl.Acad.Sci.USA,1988,85:5784-5788.
[22]Takai T,Yokoyama C,Wade K,et al.Primary structure of chichen liver acetyl-CoA carboxylase deduced from cDNA sequence [J].J.Biol.Chem.,1988,263:2651-2657.
[23]Gornicki P,Podkowinski J,Scappino L A,et al.Wheat acetyl-Coenzyme A carboxylase:cDNA and protein structure [J].Proc.Natl.Sci.USA,1993,91:6860-6864.
[24]Schulte W,Schell J,Topfer R.A gene encoding acetyl-coenzyme A carboxylase from Brassica napus[J].Plant Physiol.,1994,106:793-794.
[25]Shorrosh B S,Dixon RA,Ohlrogge J B.Molecular cloning,characterization,and elicitation of acetyl-CoA carboxylase from alfalfa [J].Proc.Natl.Acad.Sci.USA,1994,91:4323-4327.
[26]Sasaki Y,Hakamada K,Suama Y,et al.Chloroplast-encoded protein as a subunit of acetyl-CoA carboxylase in pea plant [J].J.Biol.Chem.,1993,268(33):25118-25123.
[27]Kozaki A,Mayumi K,Sasaki Y.Thiol-Disulfide exchange between nuclear encode and chloroplast-encode subunits of pea acetyl-CoA carboxylase[J].The Journal of Biological Chemistry,2001,276(43):39919-3992.
[28]Thelen J J,Mekhedov S,Ohlrogge J B.Brassicaceae express multiple isoforms of biotin carboxyl carrier protein in a tissue-specific manner[J].Plant Physiol.,2001,125:2016-2028.
[29]Polakis S E,Guchhait R B,Zwergel E E,et al.Acetyl coenzyme A carboxylase system of Eshcherichia coli,studies on the mechanisms of the biotin carboxylase and carboxyltransferase catalyxed reactions[J].The Journal Biological Chemistry,1974,249(20):6657-6667.
[30]Podkowinski J,Jelenska J,Sirikhachornkit A,et al.Expression of cytosolic and plastid acetyl-coenzyme A carboxylase genes in young wheat plants[J].Plant Physiology,2003,131:763–772.
[31]Gornichi P,Faris J,King I,et al.Plastid-localized acetyl-CoA carboxylase of bread wheat isencoded by a single gene on each of the three ancestral chromosome sets[J].Proc.Natl.Acad.Sci.USA,1997,94:14179-14184.
[32]Podkowinski J,Sroga G E,Haselkorn R,et al.Structure of a gene encoding acytosolic acetyl-CoA carboxylase of hexaploid wheat[J].Proc.Natl.Acad.Sci.USA,1996,93:1870-1874.
[33]Elborough K M,Winz R,Deka R K,et al.Biotin carboxyl carrier protein and carboxyltransferase subunits of the mutil-subunit from Brassica napus:cloning and analysis of expression during oilseed rape embryogenesis[J].Biochem.J.,1996,315:103-112.
[34]Schulte W,T?pfer R,Stracke R,et al.Multi-functional acetyl-CoA carboxylase from Brassica napus is encoded by a multi-gene family:indication for plastidic localization of at least oneisoform[J].Proc.Natl.Acad.Sci.USA,1997,94(7):3465-3470.
[35]Kimura Y,Miyake R,Tokumasu Y,et al.Molecular cloning and characterization of two genes for the biotin carboxylase and carboxyltransferase subunits of acetyl coenzyme A carboxylase in Myxococcus xanthus[J].Journal of Bateriology,2000,182 (19):5462-5469.
[36]Abu-Elheiga L,Matzuk M M,Kordari P,et al.Mutant mice lacking acetyl-CoA carboxylase 1 are embryonically lethal[J].PNAS,2005,102(34):12011-12016.
[37]Abu-Elheiga L,Almarza-Ortega D B,Baldini A,et al.Human acetyl-CoA carboxylase 2 molecular cloning,characterization,chromosomal map,and evidence for two isoforms [J].The Journal of Biological Chemistry,1997,272 (16):10669-10677.
[38]Mao J,Chirala S S,Wakil S J.Human acetyl-CoA carboxylase 1 gene:Presence of three promoters and heterogeneity at the 5'-untranslated mRNA region[J].PNAS,2003,100(13):7515-7520.
[39]Widmer J,Fassihi K S,Schlichter S C,et,al.Identification of a second human acetyl-CoA carboxylase gene[J].Biochem.J.,1996,316:915-922.
[40]Post-Beittenmiller D,Roughan P G,Ohlrogge J B.In vivo pools of free and acytel acyl carrier proteins in spinach:Evidence for sites of regulation of fatty acid biosynthesis[J].Biol.Chem.,1991,266:1858-1865.
[41]Roughan P G.Stromal concentrations of coenzyme A and its esters are insufficient to account for substrate channelling within the chloroplast fatty acid synthase[J].Biochem.,1997,327:267-273.
[42]李亮,程彥伟.乙酰辅酶A羧化酶在治疗肥胖中的潜在作用[J].生命的化学,2007,27(2):180-182.
[43]Tong L.Acetyl-coenzyme a carboxylase:crucial metabolic enzyme and attractive target for drug discovery[J].Cell Mol.Life Sci.,2005,10:1007-1018.
[44]Roesler K,Shintani D,Savage L,et al.Targeting of the Arabidopsis homomeric acetyl-coenzyme A carboxylase to plastids of rape seeds [J].Plant Physiol.,1997,113:75-81.
[45]Shintani D K,Ohlergge J B.Feedback inhibition of fatty acid synthesis in tobacco suspension cells[J].The Plant Journal,1995,7(4):577-587.
[46]姜莉莉,史晓斌.ACCase抑制剂类除草剂的作用机理[J].农药研究与应用,2010,4:14-17.
[47]衣克寒,付穎,叶非,等.乙酰辅酶A羧化酶抑制剂的构效关系和抗性研究进展[J].植物保护,2012,38(1):11-17.
[48]Deng W,Cai J,Zhang J,et al.Molecular basis of resistance to ACCase-inhibiting herbicide cyhalofop-butyl in Chinese sprangletop (Leptochloa chinensis (L.) Nees) from China.Pesticide Biochemistry and Physiology,2019,158:143-148.
[49]Liu B,Ding F,Wang M ,et al.Cross-resistance pattern to ACCase-inhibiting herbicides in a novel Trp1999Leu mutation American sloughgrass (Beckmannia syzigachne) population[J].Pesticide Biochemistry and Physiology,2019,159:80-84.
[50]Reverdatto S,Beilinson V,Nielsen N C,et al.A multisubunit acetylcoenzyme A carboxylase from soybean[J].Plant Physiology,1999,119:961-978.
[51]Ke J,Wen T N,Nikolau B J,et al.Coordinate regulation of the nuclear and plastidic genes coding for the cubunits of the heteromeric acetyl-coenzyme A carboxylase[J].Plant Physiology,2000,122:1057-1071.
[52]Li Shyr-Jiann,Cronan J E.Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase,which catalyzes the first committed step of lipid biosynthesis[J].Journal of Bacteriology,1993,175(2):332-340.
[53]Hedtke B,B?rner T,Weihe A.Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis[J].Science,1997,277:809-811.
[54]Maliga P.Two plastid RNA polymerases of higher plants:an evolving story[J].Trends Plant Sci.,1998,3:4-6.
[55]Swiatecka-Hagenbruch M,Liere K,Borner T.High diversity of plastidial promoters in Arabidopsis thaliana[J].Mol.Genet.Genomics,2007,277:725-734.
[56]Madoka Y,Tomizawa K,Mizoi J,et al.Chloroplast transformation with modified accD operon increases acetyl CoA carboxylase and causes extension of leaf longevity and increase in seed yield in tobacco[J].Plant Cell Physiol.,2002,43:1518-1525.
[57]Shinozaki K,Ohme M,Tanaka M,et al.The complete nucleotide sequence of the tobacco chloroplast genome:its gene organization and expression [J].EMBO J.,1986,5:2043-2049.
[58]Nagano Y,Matsuno R,Sasaki Y.Sequence and transcription analysis of the gene cluster trnQ-zfpA-psaI-ORF231-petA in pea chloroplasts[J].Curr.Genet.,1991,20:431-436.
[59]Hajdukiewicz P T J,Allison L A,Maliga P.The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids [J].EMBO J.,199716:4041-4048.
[60]Hirata N,Yonekura D,Yanagisawa S,et al.Possible involvement of the 5'-flanking region and the 5'UTR of plastid accD gene in NEP-dependent transcription[J].Plant Cell Physiol.,2004,45(2):176-186.
[61]Sasaki Y,Kozaki A,Ohmori A,et al.Chloroplast RNA editing required for functional acetyl-CoA carboryase in plants [J].The Journal of Biological Chemistry,2001,276(6):3937-3940.
[62]Sugiura M,Hirose T,Sugita M.Evolution and mechanism of translation in chloroplasts[J].Annu.Rev.Genet.,1998,32:437-459.
[63]Hou B K,Zhou Y H,Wang L H,et al.Chloroplast transformation in oilseed rape[J].Tansgenic Res.,2003,12:111-114.
[64]Shintani D,Roesler K,Shorrosh B,et al.Antisense expression and overexpression of biotin carboxylase in tobacco leaves[J].Plant Physiol.,1997,114(3):881-886.
[65]Roesler K R,Schorrosh B S,Ohlrogge J B.Structure and expression of an Arabidopsis acetyl-coenzyme A carboxylase gene [J].Plant Physiol.,1994,105:611-617.
[66]Sasaki Y,Konishi T,Nagano Y.The compartmentation of acetyl coenzyme A carboxylase in plants[J].Plant Physiol.,1995,108:445-449.
[67]楚敏,赵虎基,郑明刚,等.谷子乙酰辅酶A羧化酶BC功能域的克隆及原核表达载体的构建[J].植物生理学报,2004,22(5):408-410.
[68]Lee S S,Jeong W J,Bae J M,et al.Characterization of the plastid-encodeed carboxyltransferase subunit (accD) gene of potato [J].Mol.Cell,2004,17(3):422-429.
[69]Herbert D,Price L J,Alban C,et al.Kinetic studies on two isoforms of acetyl-CoA carboxylase from maize leaves[J].Biochem.J.,1996,318:997-1006.
[70]Li Z G,Yin W B,Guo H,et al.Genes encoding the alpha-carboxyltransferase subunit of acetyl-CoA carboxylase from Brassica napus and parental species:cloning,expression patterns,and evolution[J].Genome,2010,53(5):360-370.
[71]武玉永,譚秀华,马立新.甘蓝型油菜乙酰辅酶A羧化酶3个亚基的克隆及其表达[J].安徽农业科学,2008,36(10):4002-4006.
[72]Li M,Xia H,Zhao C,et al.Isolation and characterization of putative acetyl-coA carboxylases in Arachis hypogaea L.[J].Plant Mol.Biol.Rep.,2010,28:58-68.
[73]Qiao Z and Liu J.Cloning and characterization of cotton heteromeric acetyl-CoA carboxylase genes[J].Progress in Natural Science,2007,17(12):1412-1418.
[74]Nakkaew A, Chotigeat W, Eksomtramage T,et al.Cloning and expression of a plastid-encoded subunit,beta-carboxyltransferase gene(accD) and a nuclear-encoded subunit,biotin carboxylase of acetyl-CoA carboxylase from oil palm (Elaeis guineensis Jacq.) [J].Plant Science,2008,175(4):497-504.
[75]Gu K,Chiam H,Tian D,et al.Molecular cloning and expression of heteromeric ACCase subunit genes from Jatropha curcas[J].Plant Sci.,2011,180 (4):642-649.
[76]武玉永,马立新,蒋思婧.甘蓝型油菜羧基转移酶A亚基全长cDNA的克隆及在大肠杆菌中表达[J].生物化学与生物物理进展,2004,31(9):847-854.
[77]卢捷,姚玉峰,姜卫红,等.地中海拟无枝菌酸菌U32中生物素羧基载体蛋白结构基因的克隆、表达及转录[J].微生物学报,2003,43(1):56-64.
[78]李孟軍,夏晗,王兴军,等.花生野生近缘种生物素羧基载体蛋白基因的克隆与结构分析[J].华北农学报,2009,24(6):6-10.
[79]Cui Y,Liu Z,Zhao Y,et al..Overexpression of heteromeric GhACCase subunits enhanced oil accumulation in Upland cotton.Plant Mol.Biol.Rep.,2017,35:287-297.
[80]Atsushi Inamura.Yayoi Ohashi,Etsuko Sato,et al.Intraspecific sequence variation of chloroplast DNA reflecting variety and geographical distribution of Polygonurn cuspidatum(Polygonaceae) in Japan[J].J.Plant Res.,2000,113:419-426.
[81]Matsuda Yuji,Yoshimura Hitoshi,KanamotoHirosuke,et al.Sequence variation in the rbcL-accD region in the chloroplast genome of Moraceae[J].Plant Biotechnology,2005,22(3):231-233.
[82]Alrefai R,Berke T G,Rocheford T R.Quantitative trait locus analysis of fatty acid concentrations in maize[J].Genome,1995,38:894-901.
[83]Kianian S F,Egli M A,Phillips R L,et al.Association of major groat oil content QTL and an acetyl-CoA carboxylase gene in oat[J].Theor.Appl.Genet.,1999,98:884-894.
[84]Yang X,Guo Y,Yan J,et al.Major and minor QTL and epistasis contribute to fat compositions and oil concentration in high-oil maize[J].Theor.Appl.Genet.,2010,120:665-678.
[85]陈江林,幸伟年,唐佰平.油桐种仁不同发育时期ACCase活性与含油率相关性分析[J].南方林业科学,2016,44(5):14-16,34.
[86]王保明.油茶ACCase基因的克隆及功能研究[D].长沙:中南林业科技大学,2012.
[87]Wang B,Tan X,Jiang J,et al.Molecular cloning and expression of two genes encoding ACCase subunits of Camellia oleifera (Theaceae)[J]. Pak.J.Bot.,2018,50(1):103-110.
[88]王哲,油桐异质型ACCase基因的克隆及功能表达研究[D].长沙:中南林业科技大学,2015.
[89]Cui Y,Zhao Y,Wang Y,et al.Genome-wide identification and expression analysis of the biotin carboxyl carrier subunits of heteromeric acetyl-CoA carboxylase in Gossypium[J].Front.Plant Sci.,2017,8:624.
[90]崔宇鹏.棉花异质型ACCase基因家族鉴定与功能分析[D].北京:中国农业大学,2017.
[91]Kozaki A,Kamado K,Nagano Y,et al.Recombinant carboxyltransferase responsive to redox of pea plastidic acetyl-CoA carboxylase [J].Biol.Chem.,2000,275(14):10702-10708.
[92]Kahlau Sand Bock R.Plastid transcriptomics and translatomics of tomato fruit development and chloroplast- to-chromoplast differentiation:chromoplast gene expression largely serves the production of a single protein[J].Plant Cell,2008,20:856-874.
[93]Davias M S,Solbianti J,Cronan J E.Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherich coli[J].Biol.Chem.,2000,275(37):28593-28598.
[94]Zha W, Rubin-Pitel S B, Shao Z,et al.Improving cellular malonyl-CoA level in Escherichia coli via metabolic engineering[J].Metabolic Engineering,2009,11(3):192-198.
[95]Ohlrogge J B,Roesler K R and Shorrosh B S.Methods of increasing oil content of seeds[P].United States Patent:5925805,1999-7-20.
[96]Klaus,D,Ohlrogge,J B,Neuhaus H E,et al.Increased fatty acid production in potato by engineering of acetyl-CoA carboxylase[J].Planta,2004,219:389-396.
[97]Sellwood C,Slabas A R,Raw sthorne S.Effects of manipulating expression of acetyl-CoA carboxylase in Brassica napus L.embryos[J].Biochemical Society,2000,28:598-600.
[98]Skarjinskaia M,Svab Z,and Maliga P.Plastid transformation in Lesquerella fendleri,an oilseed Brassicacea[J].Transgenic Res.,2003,12:115-122.
[99]Kode V,Mudd E A,Iamtham S,et al.The tobacco plastid accD gene is essential and is required for leaf development [J].Plant J.,2005,44 (2):237-244.
[100]Thelen J J and Ohlrogge J B. Both antisense and sense expression of biotin carboxyl carrier protein isoform 2 inactivates the plastid acetyl-coenzyme A carboxylase in Arabidopsis thaliana[J]. Plant. J., 2002, 32:419-431. (責编:张宏民)
