In situ Assemblies of Bacteria and Nutrient Dynamics in Response to an Ecosystem Engineer, Marine Clam Scapharca subcrenata, in the Sediment of an Aquaculture Bioremediation System
2020-11-30NICHOLAUSReganLUKWAMBEBetinaYANGWenZHUJinyongandZHENGZhongming
NICHOLAUS Regan, LUKWAMBE Betina, YANG Wen, ZHU Jinyong, and ZHENG Zhongming, *
Assemblies of Bacteria and Nutrient Dynamics in Response to an Ecosystem Engineer, Marine Clam, in the Sediment of an Aquaculture Bioremediation System
NICHOLAUS Regan1), 2), LUKWAMBE Betina1), 3), YANG Wen1), ZHU Jinyong1), and ZHENG Zhongming1), *
1)School of Marine Sciences, Ningbo University, Ningbo 315832, China 2) Department of Natural Sciences, Mbeya University of Science and Technology, Mbeya 53000, Tanzania 3)Department of Food Science and Technology, University of Dar es Salaam, Dar es Salaam 35091, Tanzania
The relationships between nutrient dynamics, microbial community, and macrofauna are important in bioremediation systems. In this study, we examined the effects of marine clamsedimentary activities on the microbial assemblages, benthic nutrient chemistry, and its subsequent remediation impacts on organic effluent in the sediment of an aquaculture wastewater bioremediation system. The results showed that microbial community composition differed significantly in the clam area (ANOSIM,=0.707,=0.037). Pyrosequencing of bacterial 16S rRNA gene revealed a total of 48 unique phyla, 79 classes, 107 orders, 197 families, and 321 genera amongst all samples. The most dominant bacterial assemblages were Proteobacteria, Bacteroidetes, Acidobacteria, Firmicutes, Verrucomicrobia, and Actinobacteria, with Bacteroidetes and Firmicutes significantly higher in all treatment samples than control (<0.001). All dominant phyla in the list were shared across all samples and accounted for 89% (control) and 97% (treatment) of the total 16S rRNA. The nutrient flux rates from the sediments into the water (treatment group) were 51% (ammonium), 88% (nitrate), 77% (nitrite) and 45% (phosphate) higher, relative to the control implying increased mineralization, degradability, and mobility of the benthic nutrients. Similarly, significantly increased oxygen consumption rates were evident in the clam area signifying improved oxygen distribution within the sediment. The organic effluent contents associated with total- organic matter, carbon, nitrogen, and, phosphate were lower among the clam treatments relative to the control. Our results describe the potential roles and mechanisms contributed by marine bivalveon benthic-bacterial-community assembly, nutrient balance, and effluent reduction in the sediments of aquaculture wastewaters bioremediation system.
; effluents; bioturbation: sediment chemistry; bioremediation; nutrient dynamics
1 Introduction
Intensive aquaculture has a significant adverse environmental impact due to the release of great amounts of organic wastes (Karakassis., 2000). For instance, wastewater generated from aquaculture productions is normally characterized by high chemical effluents associated with nitrogen wastes (ammonia, nitrate, and nitrite; Dauda., 2014) as well as uneaten feeds and excretion products. These products usually pollute the ecosystem of the receiving water bodies causing severe alterations of the physical-chemical properties and eventually accumulates in the sediments (Diaz, 2001). This understanding has introduced many apprehensions about the environmental impacts of aquaculture wastes. One of the key challenges for the sustainable development of aquaculture is the reduction of its environmental impacts. To maintain sustainable aquaculture production and its subsequent environmental pollution, various effluent remediation methods have been established including integrated aquaculture effluent bioremediation systems (Brito., 2018; Lukwambe., 2018).
Integrated aquaculture effluent bioremediation systems involving macrofauna such as bivalves is one of the most cost-effective and rising biological wastewater treatment technology (Lukwambe., 2018; Nicholaus., 2019b). The effectiveness of this system largely depends on the interaction between the sediment microbial community, and the benthic macrofauna activities (., clam- burrowing) (Lukwambe., 2018; Nicholaus., 2019b). The practice of incorporating clam in aquaculture restoration systems to mitigate for the excess nutrients is common and predicated on the idea that clam will optimize and/or enhance the microbial growth and activities, and nutrient cycling in the system. Clams are the most important component in the system. They could not only reduce the nutrients by feeding on organic detritus but also remediate the sediment during the bioturbation process (., Nicholaus., 2019a, b). For example, the organic carbon produced by fish farming in the form of uneaten feed and excretions eventually deposit in the sediment and represent a great source of food for filter-feeding organisms, such as bivalves (Navarrete-Mier., 2010). Similarly, clams among other macrofauna inhabiting marine sediments markedly alter the physical structure and chemical composition and microbial biomass turnover (., Sinkko., 2011; Shen., 2017). In this case, clams played concurrent activities (feeding, bioirrigation, digging, mineralization, and burrowing) which are commonly referred to as ‘bioturbation’, a process of sediment reworkings by benthic animals (Aller, 2001). This process offers a significant ecological role in bioremediation practices (Nicholaus., 2019b).
In the integrated model, sediments are responsible for transporting and cycling of a significant proportion of many nutrients and contaminants (Zhao., 2019). It is estimated that up to 80% of the nitrogen needed by the primary producers in the aquatic system is provided by sediment remineralization reactions (Dale and Prego, 2002). Conversely, marine sediments represent an important dynamic of inorganic nutrientsanaerobic decompositions regulated by microorganism under influence of bioturbating organisms (Lukwambe., 2018). Similarly, microorganisms are very useful and with major importance in aquaculture industrial wastewater treatment systems. They dictate several positive effects on the fate of aquaculture operations (Moriarty, 1997). The greatly microbial roles include the elimination of toxic environmental materials (ammonia, nitrite, and hydrogen sulfide), degradation of uneaten feed, and management of nutrient cycling and water quality (Moriarty, 1997; Yang., 2018). These among other functions make the interaction between clams and microorganisms key players in aquaculture bioremediation systems.
Various studies including the commonly known ecosystem engineers like filter-feeder bivalves (Mussels: Zhang., 2011; Razor clam: Lukwambe., 2018; Zhao., 2019; blood cockles: Nicholaus., 2019b) have been previously reported to significantly influence nutrient dynamics and improve bioremediation environment. Moreover, to improve the previous studies, and collect sufficient reports that describe the unique roles and mechanisms played by intertidal shall burrower marine bivalves such as(, Zhang., 2020) in marine ecological sediment-based bioremediation system is necessary. Therefore, the present study aimed to 1) investigate and characterize the distribution and assembly of the microbial community in a field ecological bivalve- sediment based bioremediation system, 2) examine the ecosystem bioengineering mechanisms ofin boosting sediment microorganism, and 3) examine its potential effects on benthic biogeochemical nutrient fluxes and organic wastes mitigation in a field ecological aquaculture wastewater treatment system. This study will enlighten the role played by marine clam.in regulating the benthic microbial community assembly, nutrient dynamics, and organic effluent reduction in search of an optimized clam-sediment-bioengineering based bioremediation system, a recently rising and viable approach for aquaculture-wastes and contaminants management approach.
2 Material and Methods
2.1 Study Area and Experimental Design
The ecological aquaculture wastewater treatment system studied is situated in Xiangshan Bay, in Ningbo, China. This project was built in 2017 purposely to improving and optimizing the aquaculture wastewater treatments. The studied system consists of 4 effluent treatment sections arranged serially: 1) Physical sedimentation; 2) Biofilms; 3) Bivalves; 4) Artificial wetlands (Lukwambe., 2018). The biofilters species (., bivalves and wetlands) were changed and the performance of the system was compared in different production seasons to get the optimized system. In this study, the clam cultured zone: about 800m2(2/5 of the total area) was chosen as the treatment area. And an area between biofilm and bivalve (clam) was chosen as the ‘control’. Eight partitions/enclosures, separated by PVC fabric mesh (100% polyester) were formed, 4 for control, and the other 4 for the treatment. Before the experiment, the bottoms of each enclosure were unified in order that they contain a similar initial condition. High vitality clams () with an average length of 1.8±0.3cm were stocked at an average stocking density of 360indm−2(Zhang., 2020). The average depth of each treatment enclosure was approximately 1.2m. and the size of the shrimp farm in which the effluents were produced is approximately 3300m2. The wastewater from the intensive aquaculture shrimp () ponds to the treatment system was discharged daily in the morning between 7:00–9:00 am. More than 165m3of effluent water was discharged daily. The average nutrients removal efficiency of the system is, 81.64% (total nitrogen (TN)), 82.34% (total phosphate (TP)), 75.68% (chemical oxygen demand), and 72.18% (ammonium (NH4+-N)) (Lukwambe., 2019b).
2.2 Sample Collections
2.2.1 Sediment microbial and biogeochemical samples
During sampling, a total of 8 undisturbed sediment cores (approximately 8–9cm each), were collected using a handheld coring device (1.6m) with a removable round plexiglass tube (11cm i.d., 45cm height). Four for the control and four for the treatment to investigate 1) benthic biogeochemical nutrient fluxes, 2) sediment contents, and 3) the 16S rRNA microbial community assemblages. After sampling, the sediment cores (in the polypropylene tubes) and about 40L of thewater were brought back to the laboratory within 1h of collection. The plexiglass tubes were adapted with an injection port at 2cm above the sediment grind for drawing nutrient-flux water samples.
2.2.2 Water column characteristics
The dissolved oxygen (DO), temperature, salinity, and pH profiles of the water column were determinedusing a portable field multiparameter probe (YSI-550A oxygen meter, USA). Water column samples for phosphate (PO43−-P), nitrate (NO3−-N), nitrite (NO2−-N), and NH4+-N were also sampled for the determination of nutrient levels in the system.
2.3 Biogeochemical Nutrient Incubation, Sampling and Flux Analytics
The biogeochemical nutrient flux sampling and measurement procedures were carried as previously described (Zheng., 2009; Nicholaus and Zheng, 2014). Briefly, the sediment core tubes were slowly filled with fresh- water (from the site), and sealed with plastic stopper then incubated in water baths for 3h. Then rates of nutrient exchange were measured by extracting 45mL water samples with a plastic syringe at an interval of 1h through a hole bored across the tube at 1cm above the sediment surface. The concentration of collected nutrient samples was corrected for the composition of the water used for the replacement of extracted sample at each sampling time (Zheng., 2009). Extracted water samples for NH4+-N, NO2−-N, NO3−-N, and PO43−-P were collected in a screw cap polypropylene bottles, filtered (0.45μm cellulose acetate filter, GF) and immediately stored frozen (−20℃) until further analysis. To estimate the sediment oxygen flux, the DO level of the overlying water in each sediment core was measured at the beginning (0h) and the end (3h) of the incubation experiment using YSI- 550A oxygen meter (USA). Then all water samples for nutrient analyses were determined calorimetrically using a WESTCO SmartChem discrete analyzer (USA). The NO2−-N and NO3−-N were measured with the cadmium- copper reduction method. NH4+-N was measured by the indophenol blue method and the PO43−-P flux concentration was determined using the ammonium molybdate/ ascorbic acid method. The net biogeochemical nutrients and DO fluxes rates were calculated from the slopes of a linear regression concentration against time equation:
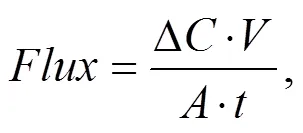
where:(µmolm−2h−1) is the sediment O2or nutrients flux;(mgL−1) is the change in concentration of oxygen or nutrients (prior and after incubation);(m3) is the volume of the overlying water;(m2), is the cross-sectional area of the incubation chamber;(h) is the duration of incubation.
2.4 Sediment TP, TOC, TN and OM
The sediment samples were oven-dried at 80℃ to constant mass. The dried samples were ground to pass through a 1-mm mesh size before analyses. The analyses included the proportion of organic matter (OM) which was measured as a percentage loss upon ignition, LOI% for 4h at 550℃ after conversion of carbon. The TP contents were measured as previously described (Mudrich, 1996) while TN and total organic carbon (TOC) concentrations were determined by an Elemental NC Analyzer (Germany) after HCl acidification of the sediment samples to remove carbonate.
2.5 Extraction and Sequencing of the Sediment Microbial DNA
Post the nutrient flux incubation experiment, the intact sediment cores were sectioned at a different interval to make three cores samples: 0–2cm (surface), 2–4cm (middle), and 4–8cm (bottom). Before extraction, the sediment samples were freeze-dried by freeze dryer (Germany). Later, the genomic sediment DNA was extracted in triplicate from approximately 0.5g sediment of each replicate sample by using the manufacturer’s instructions given in the Power Soil DNA Isolation Kit (MoBio, USA). This process yielded a purified genomic sediment DNA for use in polymerase chain reaction (PCR) amplification. The extraction efficiency was improved by a disruptor (Bio101) FastPrep cell. DNA from the triplicate samples was pooled into a final volume of 50µL in Tris-EDTA buffer. DNA concentrations (quality and quantity) were quantified spectrophotometrically using an ND-2000 Nanodrop spectrometer (Thermo Fisher Scientific, Wilmington, DE) at 260nm absorption and 260/280 and 260/230nm absorbance ratios.
High-throughput 16S rRNA gene sequencing was performed on the Illumina MiSeq platform (Illumina, San Diego, CA, USA). The V3–V4 hypervariable regions of the 16S rRNA gene were amplified by pyrosequencing to analyze the microbial community (composition, structure, and diversity) using fusion primers consisting of sequencer specific nucleotides, multiplex identifier tag, and template-specific nucleotide with the template specific sequences within the forward 341F (5’-CCTAYGGGRBG CASCAG-3’) and reverse 806R (5’-GGACTACNNG GGTATCTAAT-3’) barcoded PCR primers in 10ng DNA template. The V3-V4 hypervariable regions of the 16S ribosomal ribonucleic acid (rRNA) gene was amplified in triplicate PCR reactions (5μL volume in 50-μL reaction system). Briefly, according to Nicholaus. (2019b) the PCR cycle conditions for the 16S rRNA segment gene for each sample was carried out as follows: denaturation at 95℃ for 3min, and 27 cycles at 95℃ for 30s, 55℃ for 30s, 72℃ for 45s, plus a final extension of 10min at 72℃. Triplicate PCR amplicons were mixed after purification using a Takara purification kit (Takara, Japan). Libraries were then generated using TruSeq™ DNA Sample Prep Kit for Illumina (Illumina, San Diego, CA, USA), following the manufacturer’s instructions. Library construction and sequencing were performed commercially (MajorBio Co. Ltd., Beijing).
2.6 Bioinformatics Analysis
The sequencing of paired reads was first joined with FLASH using default settings (Magoč and Salzberg, 2011). Then all analyses of the V3-V4 hypervariable regions of the microbial 16S rRNA amplicons (the Raw FASTQ files) were conducted by using the QIIME software package according to Caporaso. (2010). We demultiplexed and quality filtered the Raw 454 reads by removing low quality reads. Sequences shorter than 150 bp were discarded as well as the removal of the Roche adapters, linkers, primers, and sample barcodes (Caporaso., 2010). Microbial phylotypes were denoised to reduce possible sequencing errors and clustered at 97% identity in the operational taxonomic unit (OTU) using UCLUST (Edgar, 2010). One representative sequence of each cluster was aligned to the Greengenes database (release 13.8) with PyNAST 1.1 (DeSantis., 2006). To avoid artifacts resulting from the sample size, the chimeric sequences were detected using the Chimera Slayer algorithm in the QIIME 1.8.0 software package. OTUs observed in only one sample or represented by only one sequence (singleton OTUs) were removed. Only sequences flagged as non-chimeras by de novo and reference-based (Greengenes database) methods were retained for further analyses. Finally, the number of sequences assigned to each OTU was summarized in a table generated by QIIME. The OTU table produced by the quality filtering containing the taxonomic assignment of the OTUs were used for all downstream analyses.
2.7 Statistical Analysis
2.7.1 Microbial community composition and diversity
Microbial community composition and diversity: Statistical approaches involving alpha diversity measures were calculated to provide information about the diversity’ within individual samples in all treatments (depths/sites). Sequences were normalized to the minimum number of reads per sample and alpha diversity parameters were computed using QIEME 1.8.0 (Caporaso., 2010). Community richness estimators; Chaol and Shannon diversity were estimated. Non-metric multidimensional scaling-NMDS, (metaMDS function, based on Bray-Curtis distance matrix) were adopted to visualize the dynamics of bacteria assemblage structure (Hallett., 2016). Before NMDS, Hellinger transformations were applied to microbial data. By using the function ‘adonis’ in the vegan package, the differences in bacterial communities among the sampling points were tested with permutational multivariate analysis of variance (PERMANOVA; Anderson, 2001).
2.7.2 Microbial biomarker discovery and visualization
The differentially significant taxonomic biomarkers/ features of the control and clam areas were identified by submitting the OTU lists of samples to the LEfSe (Linear discriminant analysis Effect Size) pipeline (LDA Effect size, http://huttenhower.sph.harvard.edu/galaxy/, Segata., 2011). This method applies robust statistical tests (Kruskal-Wallis) which involve biological consistency (Wilcoxon rank-sum tests, Segata., 2011) to predict taxonomic biomarkers based on effect-size estimation. Differentially abundant and biologically predominant taxa (phylum to genus level) were determined.
2.7.3 Sediment nutrient fluxes
A one way and 2-way analysis of variance (ANOVA) were used to test the variations in nutrient flux rates. Normal distribution and homogeneity of variance were tested for each data set, and data were log-transformed when necessary to fulfill the assumptions of ANOVA. Pearson’s correlation analysis was conducted to determine the links between the bacterial community and the environmental factors. Canonical Correspondence Analysis (CCA) was carried out to correlate the environmental variables with the sediment microbial community. Generally, all statistical analyses were performed using SPSS- software (SPSS, v16.0) and the open-source software R v.3.5.1 (R Core Team, 2017). The results were generally expressed as mean±standard error and differences were considered significant at alpha <0.05.
3 Results
3.1 Physicochemical Characteristics of the Sediment and Water
The sediment cores from the control and treatment exhibited strong physical differences. The sediment from the clam zone was loose, soft, and fluffy whereas sediment texture in the control area appeared more compact, less fluffy, and coarser (physical touch and visual observations). Moreover, thecultured zone had increased brownish coloration suggesting an oxidized se- diment surface. The sediment TN, TOC, TP, and OM contents were markedly different between the treatment and the control area (ANOVA,<0.05; % dry weight-DW, Table 1). The TN, TOC, and TP contents in the clam bioturbated area were profoundly lower (<0.05). At 0–2 cm sediment dep- th (clam zone), the organic content was most altered accounting for 46% (TN), 17% (TOC), and 13% (TP) decrease compared to the control. Similarly, organic matter (OM) was reduced by 24% (DW, 0–2cm) relative to the control group.
Lower sediment organic contents may signify an enhanced OM breakdown and transformation at the clam site. Nutrient levels in the water column between the control and the treatment area were differently distributed (Table 2).Other environmental variables such as DO, temperature, and salinity were measured. Temperature were normally 24.15±0.4℃ and salinity were 26.74ppt and the two showed similar values in both the control and the clam area. The concentration of DO fol- lowed similar trends with certain degrees of variations which were not significant among the treatments (>0.05).

Table 1 The vertical sediment total nitrogen-TN, total organic carbon-TOC, total phosphorus-TP, and organic matter-OM distributions (%mean±SD) in each treatment during the present study. The different letter denotes significant differren-ces (P<0.05, ANOVA) among the same depth of different treatment groups and between depths of the same group
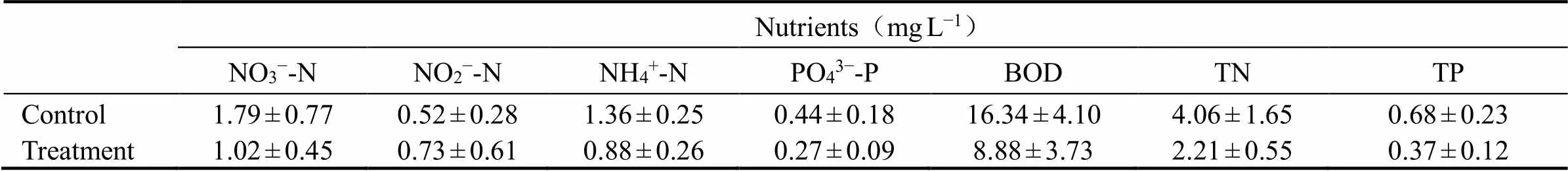
Table 2 In situ nitrate (NO3−-N), nitrite (NO2−-N), ammonium (NH4+-N), phosphate (PO43−-P), Biological oxygen demand (BOD), Total nitrogen (TN), and Total phosphate (TP) distributions in the overlying water in the studied area before the incubation experiment
3.2 Sediment-Water Fluxes of Nutrients and Sediment Oxygen Consumption (SOC)
The net nutrient fluxes between control and treatment varied significantly (Fig.1,<0.05) during the benthic nutrient flux incubation experiment. The percentage flux rate change between the treatment and the control was 51%, 88%, 77%, and 45% for NH4+-N, NO3−-N, NO2−-N, and PO43−-P respectively (Fig.1). All the nutrients showed an out-flux release trend. Concerning SOC, the clam area showed the highest consumption with mean sediment oxygen uptake of 5.07±0.52µmol O2m−2h−1(treatment) compared to 1.94±0.21µmol O2m−2h−1(control) (Fig.1). The mean rate of SOC in the sediment cores (clam area) increased significantly compared to the control group (ANOVA,<0.05). A linear regression of the oxygen concentrations showed a significant change with time (<0.05). The SOC rate (mean±SD) was roughly 3.04-fold higher than that of the control.

Fig.1 Sediment physicochemical nutrient and SOC fluxes during the incubation experiment between the control and the treatment. Data presented in µmol means±standard deviation (SD).
3.3 Microbial Community Assemblage in the S. subcrenata Sediments
Following a successful MiSeq sequencing, and after quality filtering a total of 18633 rRNA sequences representing 41584 OTUs () (clustered at 97% sequence similarity) with 48 phyla, 79 classes, 107 orders, 197 families, and 321 genera were detected amongst all samples. The number of sequences per sample ranged from 7935 to 82784, with a mean and standard deviation of 80436±26538. The highest average OTU counts (1610.91± 65.97) were obtained from the clam samples, compared to 544.39± 42.16 from the control (Table 3).

Table 3 Mean±SD values for microbial abundance and alpha diversity variation present in the sediment samples collected from the clam and control during this study
Notes: OTUs, Operational taxonomic units, Different superscript letters within the same row indicate significant differences (ANOVA;< 0.05)
Further taxonomic analysis indicated 19 unique phyla (39.6%) were significantly abundant in the clam area, 10 unique phyla (20.8%) from the control, and 19 phyla (39.6%) shared in both groups. The prominent phylain the treatments were Proteobacteria, Firmicutes, Bacteroidetes, Verrucomicrobia, Nitrospirae, Chloroflexi, Plan- ctomycete (Fig.2a, b). The most abundant bacterial assemblies (phylum level, relative abundance > 0.5% in at least one sample) were observed in clam area (surface, 0–2cm) including Proteobacteria (41.3±5.24%), Chloroflexi (33.3±2.4%), Bacteroidetes (15.7±5.26%), Firmicute (9.05±1.62%), and WS3, (4.05±1.68 %) whereas same taxa existed in the control but with slightly lower reads (Proteobacteria; 32.5±15.26%, Chloroflexi; 26.3±11.13%, Bacteroidetes; 9.4±4.22%, Firmicute; 4.37±1.68%, and WS3; 1.12±1.21%) (Fig.2a) of the total 16S rRNA seque- nces.Deeper layers, 2–4 and 4–8cm were mostly dominated by Bacteroidetes, Nitrospira, Verrucomicrobia, Ch- loroflexi, and Firmicutes in both treatments but clam samples jointly shared 76% of the total gene sequence compared to control group (22%). At class level samples were enriched with Alphaproteobacteria (29.37%), Deltaproteobacteria (24.79%), and Bacteroides (18.29%) (Fig.2b).Analysis with LEfSe further indicated the significant distribution of microbial assemblies between the clam and the control (LEfSe, LD=3.5, Fig.3). The results of the LEfSe confirmed the tendency shown in Fig.3 (phylum to genus levels) and revealed that Acidobacteria (genus of Halophageles, Candidatus, and Solibacter), Bacteroidetes (genus Flavobacterium), Pseudomonas (genus of Pseudomonadales), and Proteobacteria (genus Novosphingobium) were significantly abundant in clam area.
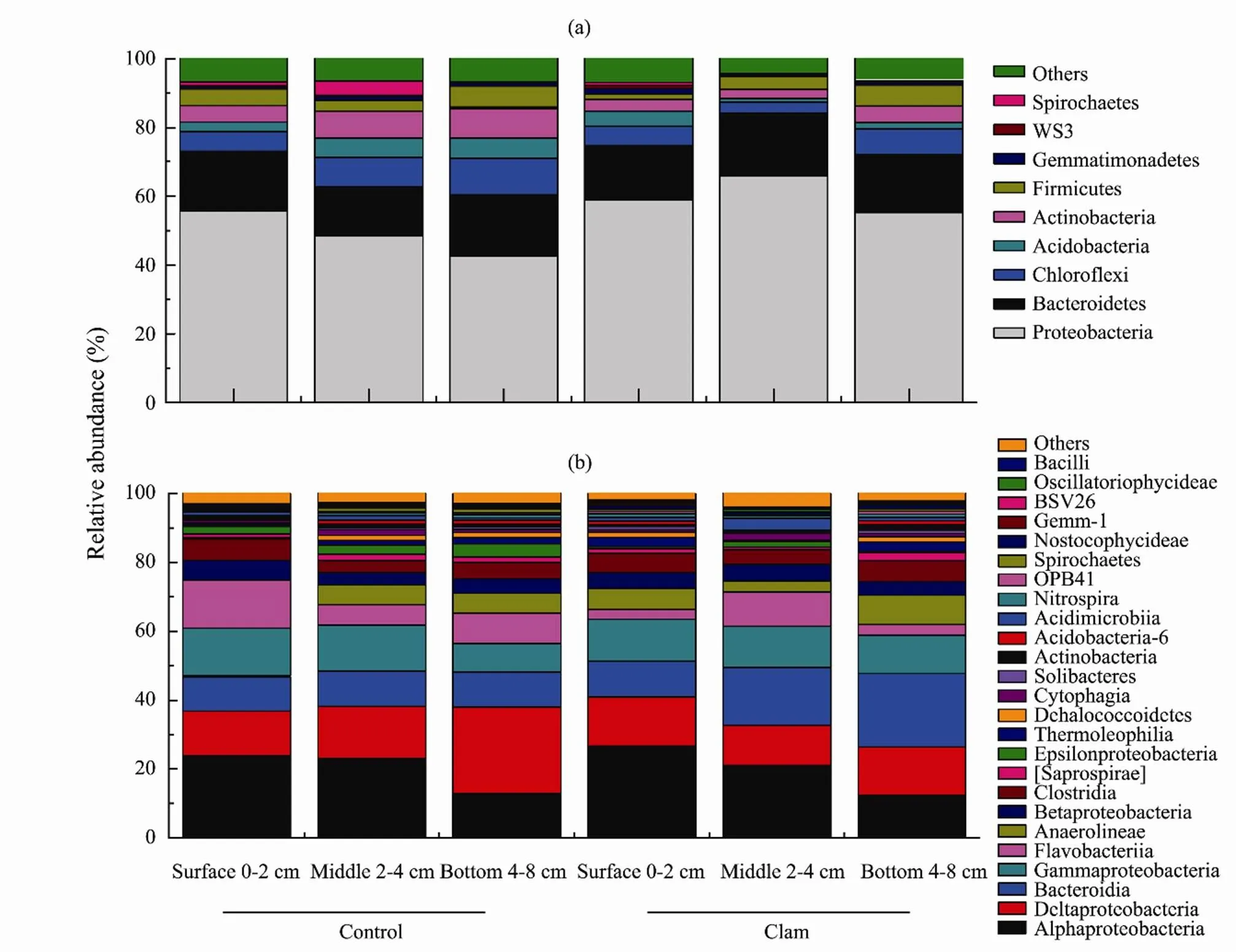
Fig.2 The relative abundance of the prominent bacterial taxa classified on phylum level (a) and (b) at class level (including Proteobacteria sub-class) from the treatments and across the core vertical depths (0–8cm). Minor phyla accounting for less than 0.5% of total sequences are summarized in the group ‘Others’.

Fig.3 The phylogenetic distribution of microbial lineages from the two study areas. Lineages with linear discriminating analysis values of 3.5 or higher determined by effect size measurements are displayed. The six rings of the cladogram stand for the domain (innermost), phylum, class, order, family, and genus. Enlarged circles in dark green and red are differentially abundant taxa identified as taxonomic biomarkers in the treatments (red=treatment, green=control).
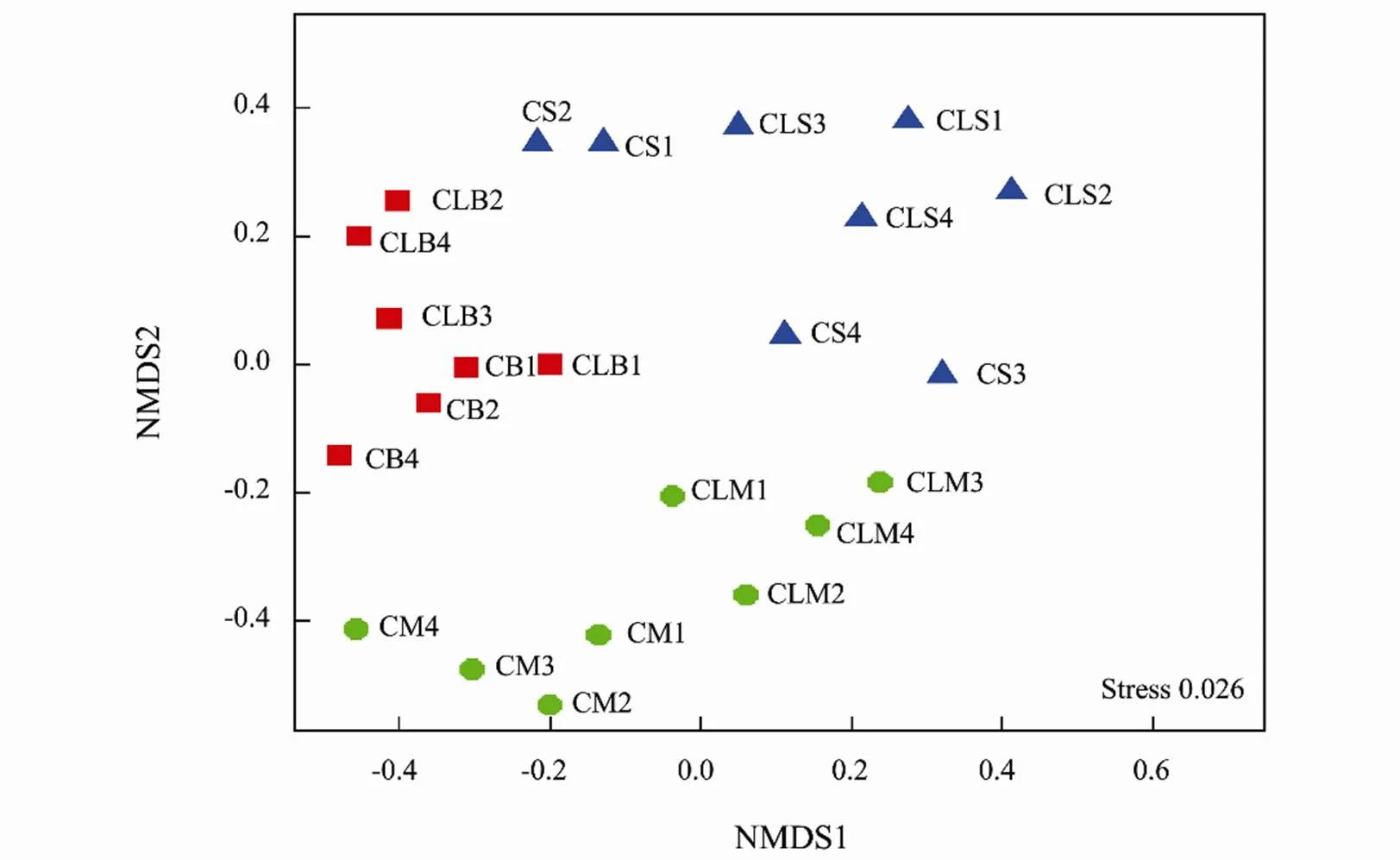
Fig.4 Non-metric multidimensional scaling (NMDS) ordination analysis plot of 16S rRNA amplicons of sediment-micro- bial-community composition of samples collected from the clam and control treatments. Community dissimilarity is based on the Bray-Curtis index. Shapes in a blue triangle, green circles, and red squares represent samples from the surface, middle and bottom respectively. Letters C and CL indicate control and clam samples respectively.
3.4 Vertical Alpha Diversity of the Bacterial Community
Bacterial diversity based on Chao1 (species richness), and Shannon (species richness and evenness), indexes of the observed OTUs varied significantly (ANOVA:<0.05, Table 3). The Simpson diversity index was non- significant (>0.05) throughout all samples, but with slightly higher reads in the clam than control samples. The Shannon index and Chao1 estimator confirmed that sediment samples at 0–2 and 2–4cm depth in the clam area experienced high microbial abundance and diversity compared to deeper depth (4–8cm) of the same treatment contrary to control group whereas no clear distribution patterns were observed. Generally, in all visualized samples, surface cores had the highest diversity indexes followed by middle and bottom cores in the clam treatment.
Analysis with NMDS ordination (Fig.4) similarly showed a considerably high diverse structural and distribution of microbial community compositions in the clam area. Taxonomically (at the order level), the surface samples had greater abundances of Methylococcales, Cytophagales, SJA-15, and Rhodobacterales, while the bottom samples were enriched with Clostridiales, Desulfobacterales, and Myxococcales. At the genera level, taxa such as Nitrospina, Pseudomonas, Planctomyces, Desulfococcus, and Verrucomicrobium were evident after the heatmap plot of the 50 most prominent genera (Fig.5). To further describe the microbial assemblages, distribution, and stru- cture we carried out the analysis of similarity (ANOSIM) and PERMANOVA test that similarly indicated marked dissimilarities among the treatments, (ANOSIM,=0.707,=0.037 and PERMANOVA,<0.05, Table 4).

Table 4 Bacterial community structure and composition determined by a permutational multivariate analysis of variance(PERMANOVA, ADONIS) for the treatment groups from the 16S rRNA sediment microbial community

Fig.5 Heatmap of differentially abundant genus taxa identified in the sediment cores, at the surface, middle, and bottom among the treatments. Rows represent centered log ratio-transformed sequence counts for each of the samples.
3.5 The Relationship Among Environmental Variables, Nutrient Fluxes, and the Bacterial Assemblies
To evaluate the relationship between the bacterial communities, nutrient fluxes, and environmental variables, CCA and PCA were conducted in the overlying-water and the sediment depth (0–2, 2–4, and 4–8cm). CCA analysis uncovered nine environmental factors (EFs) which significantly correlated with the bacterial communities and jointly accounted for 89.7% of the total variation. OM, SOC, and NO2−-N contributed the most, followed by TOC, NH4+-N, NO3−-N, TP, and PO43−-P (Fig.6a). To further explore the interlinkages between bacterial community and the EFs, PCA was drawn and the two axes jointly explain 96.1% of the microbial variance at the genus level (Fig.6b). The correlations between bacterial assemblages and EFs in both ordinations were high, indicating a strong relationship between the genus taxa and EFs. Factors such as SOC, OM, and NO2−-N were amongst the strongest correlations between the bacterial communities and the EFs. Several potential genera like,,,andwere among the taxa that highly correlated with nutrient fluxes (Figs.6a, b).
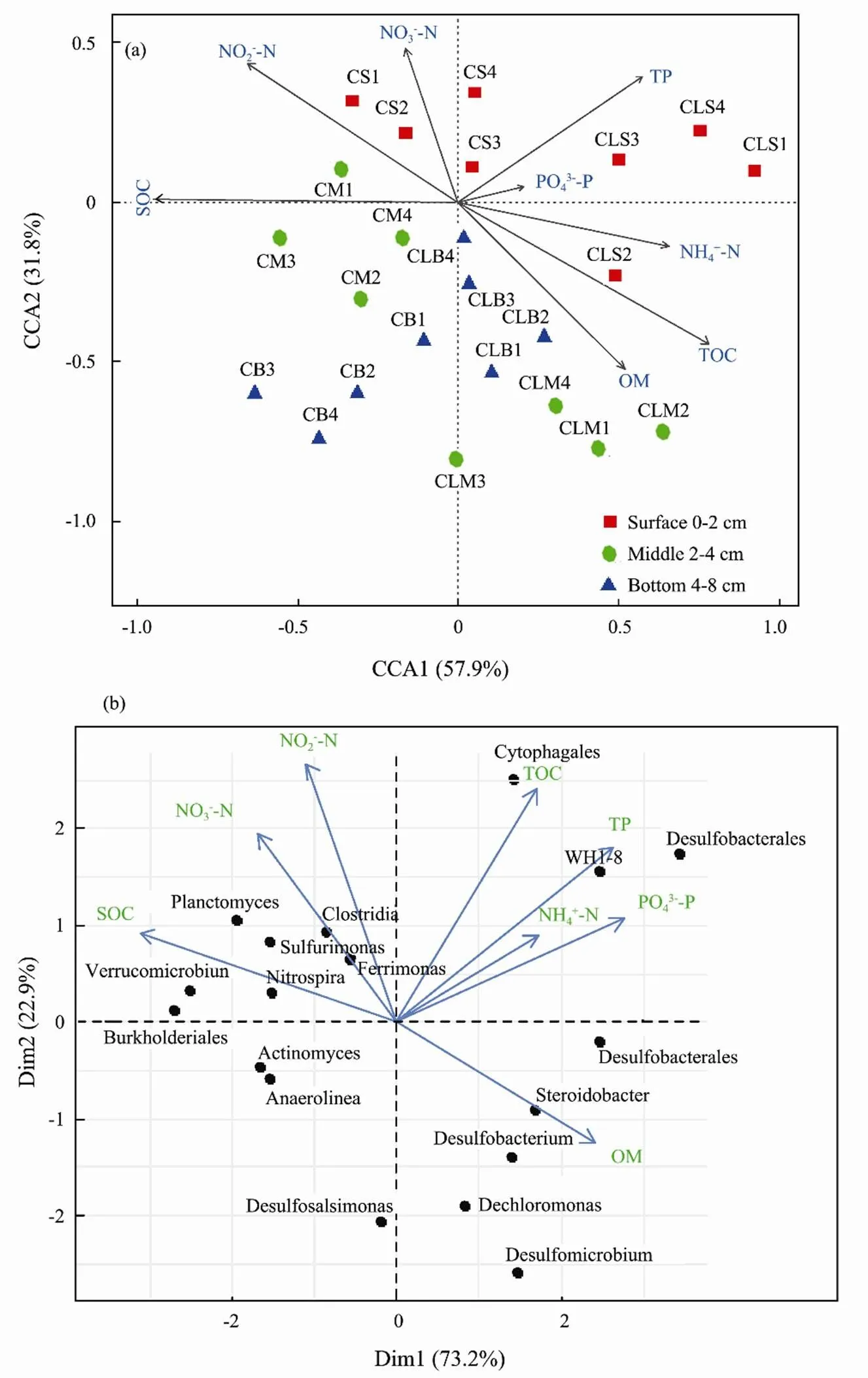
Fig.6 (a) Canonical correlation analysis (CCA) ordination plot of the microbial community composition and physicochemical variables within the treatments. (b) Principle Component Analysis (PCA) plots showing the relationship between differentially abundant genera and environmental parameters among the treatment samples during the study.
4 Discussion
Benthic macrofauna affects inorganic nutrients and microbial (processes, assemblages, and structure) in sediments due to their bioengineering activities (., Papaspyrou., 2006; Nicholaus., 2019a, b; Zhang., 2020). They are directly involved in sedimentary effluent bioremediation processes through benthic OM mineralization and aerobic metabolization (Lukwambe., 2020, Nicholaus., 2019b). These ecosystem-engi- neered roles tend to create a sustainable and ecologically balanced aquatic community. Besides, the disturbances and sediment reworking activities exerted by benthic macrofauna like bivalve filter-feeders can explain the potential mechanistic ecological process involved in microbial community assemblage, waste degradation, and the nutrient dynamics in aquatic systems (Shen., 2017; Nicholaus., 2019b).
4.1 The Impacts of Clams in the Sediment During Bioremediation Activities
The marine clamsubstantially influenced the sediment condition physiochemically and biologically. Compared to the control, the sediment in the clam area was composed of more fine particles, muddy, and silt. The organic contents associated with TN, TP, and TOC were significantly low in the clam area relative to the control (Table 1), which suggests a reduction of organic contents among the clam treatment. This finding corresponds with previous studies involving clam’s bioremediation in the sediments of aquaculture effluents (Nicho- laus., 2019b). Normallyactivities are associated with several processes such as burrowing, feeding, bioirrigation, biodeposition, resuspension, and mineralization. All these activities are believed to causing biogeochemical nutrient cycling which modifies the distribution of microorganisms responsible for bioremediation (Nicholaus., 2019b) and other microbial metabolic pathways such as sulfate reduction, sulfide oxidation, denitrification and nitrification (Kristensen and Kostka, 2005). The marine clamas a potential bioturbator induces a suitable microenvironment for OM decomposing bacteria such as(Lukwambe., 2020). Similarly, Satoh. (2007) reported a significant denitrification enhancement with ammonium oxidizing bacteria in burrows constructed by macrofauna. Microorganisms., bacteria play major roles in decomposing waste OM and removing carbonaceous compounds in the environment (Bender and Phillips, 2004). Because of the improved distribution of oxygenated water in the sediment, this may lead to increased microbial sediment OM degradations and reduction of organic effluents. These results are consistent with the previous findings which showed that the OM contents in sediments inhabited by razor clam (Lukwambe., 2018) and blood clam (Nicholaus., 2019b) were generally lower than those without
Furthermore,this study revealed substantial impacts on the benthic biogeochemical nutrient fluxes and/or cycling in the bioturbated area. Increased release of ammonium, nitrate, nitrite, and phosphate from the sediment to the overlying water was observed accounting for 51%, 88%, 77%, and 45% respectively, higher in the treatment relative to the control (Fig.1). High rates of sediment- NH4+-N and PO43−-P release due to bivalve/clam activities have been previously reported (Nicholaus., 2019a; Zhao., 2019; Zhang., 2020). The level of NH4+- N, NO2−-N, and NO3−-N concentration is linked to the changes in oxygen concentration and abundance of microorganisms observed in this study. Similarly, its inferred that the relatively higher biogeochemical nutrient fluxes in the treatment area are attributable to the nutrient mineralization activities enhanced by the clams during bioturbation. Enhanced oxygen penetration mediated via macrofaunal activities (., burrowing) is reported to increase the bioavailability of benthic elements including phosphorus fractions (Nicholaus and Zheng, 2014; Nicho- laus., 2019a). Bioturbation can widely accelerate benthic process including nutrient fluxes, nitrification rates, shift of OM to deeper sediment layers and enhanced material cycling (Nicholaus., 2020).
The SOC rates in the clam area were higher compared to that of the control (Fig.1, Table 2). The increase in oxygen influxes in the clam areas is attributable toburrowing activities in the sediments. Sediment rework by the bioturbating organism creates oxygen demand due to microbial activities such as waste degradation and mineralization. Similar results have been previously reported on the effects of clams on oxygen supply in the sediments (., Bertics and Ziebis, 2009; Ni- cholaus., 2019b). In the presence of the anaerobic condition, the process of nitrification is increased through microbial nitrate transformation which may result in a reduction of excess nitrogen and ammonium in the ecosystem.
4.2 Role of S. subcrenata on the Sedimentary Bacterial Assemblages
Macrofauna bioturbations create a fine spatial scale selective pressure that modulates significant rearrangements of sediment bacteria community assemblages and interactions (Booth., 2019). Similarly, burrows created by macrofauna can highly contain numerous potential bacteria assemblages with high abundance (Papaspyrou., 2006; Stauffert., 2014). In the present study, 48 unique bacterial phyla were widely differentiated and distributed across all samples. The clam inhabited area demonstrated a high diversity of the microbial community with significantly higher abundance of several potential bacterial phyla, such as Firmicutes, Bacteroidetes, Proteobacteria, Chloroflexi, Cyanobacteria, Lentisphaerae, Nitrospirae, Planctomycetes, Actinobacteria, Gemmatimonadetes, KSB3, and Verrucomicrobia as referred to control (Figs.2a, b). Among these taxa, the Verrucomicrobia, Chloroflexi, Firmicutes, and Planctomycetes, are one of the common taxa present in marine sediment and capable of degrading organic wastes (Lukwambe., 2018; Nicholaus., 2019b). This suggests thatimproved the sediment quality and exerted a measurable change in microbial diversity and abundance.is an intertidal clam that can potentially burrow up to 4cm deep (Nicholaus., unpublished). These burrow formations may be enriched with high oxygen-rich overlying water drawn from the surface (Bertics and Ziebis, 2009). Dissolved oxygen plus sediment median grain size, have been reported as the vital factor shaping benthic bacterial communities and OM mineralization.
Further, the distribution of some functional groups/ strains associated with sulfur-reducing bacteria in the families Desulfobacteraceae and Desulfobulbaceae (phylum: Proteobacteria, class Deltaproteobacteria) were highly represented in different OTUs, in the clam area. Again, Novosphingobium of the class Alphaproteobacterial was expressed in all samples but more explicitly in S. subcrenata area signifying that denitrification was accelerated (Feng., 2012). The resultant bivalve bioengineering activities (burrow-walls, irrigation, ventilated-holes) can alter how the microbial assemblages are structured in the sediment-ecosystem (Papaspyrou., 2006; Bertics and Ziebis, 2009). The distinct formation of microbial assemblage structures and increased diversity in the clamarea is in correspondence with other previous clam bioturbation studies (Lukwambe., 2019a, b; Nicholaus., 2019a, b). According to Ieno. (2006) and Solan. (2008), bioturbation functional intensity increased benthic species richness a process that may result in improved sediment restoration and nutrient fluxes. Mermillod-Blondin. (2004) demonstrated that, due to its strong ecosystem engineering activities, the occurrence ofincreased total active bacterial abundance in benthic sediments. Also, Chen. (2017) showed that bioturbations by macrofauna can oxygenate the sediment, affect bacterial composition, and activity.
Furthermore, the ecological process of thecan be described indirectly by the intermittent bio-irrigation of burrows that creates an oscillating oxic- anoxic ecosystem and enriching coexistence of oxic-anoxic microbial taxa (Volkenborn., 2012). The biodeposits from the clams (Fig.7) may result in high organic wastes enriched sediment which saves as food for the bacteria. Learman. (2016) demonstrated OM to be a driver of benthic microbial community structure across the surface sediment. More recently, Nicholaus. (2019b), suggested that nutrient cycling can be an important factor in altering the microorganism assemblies.
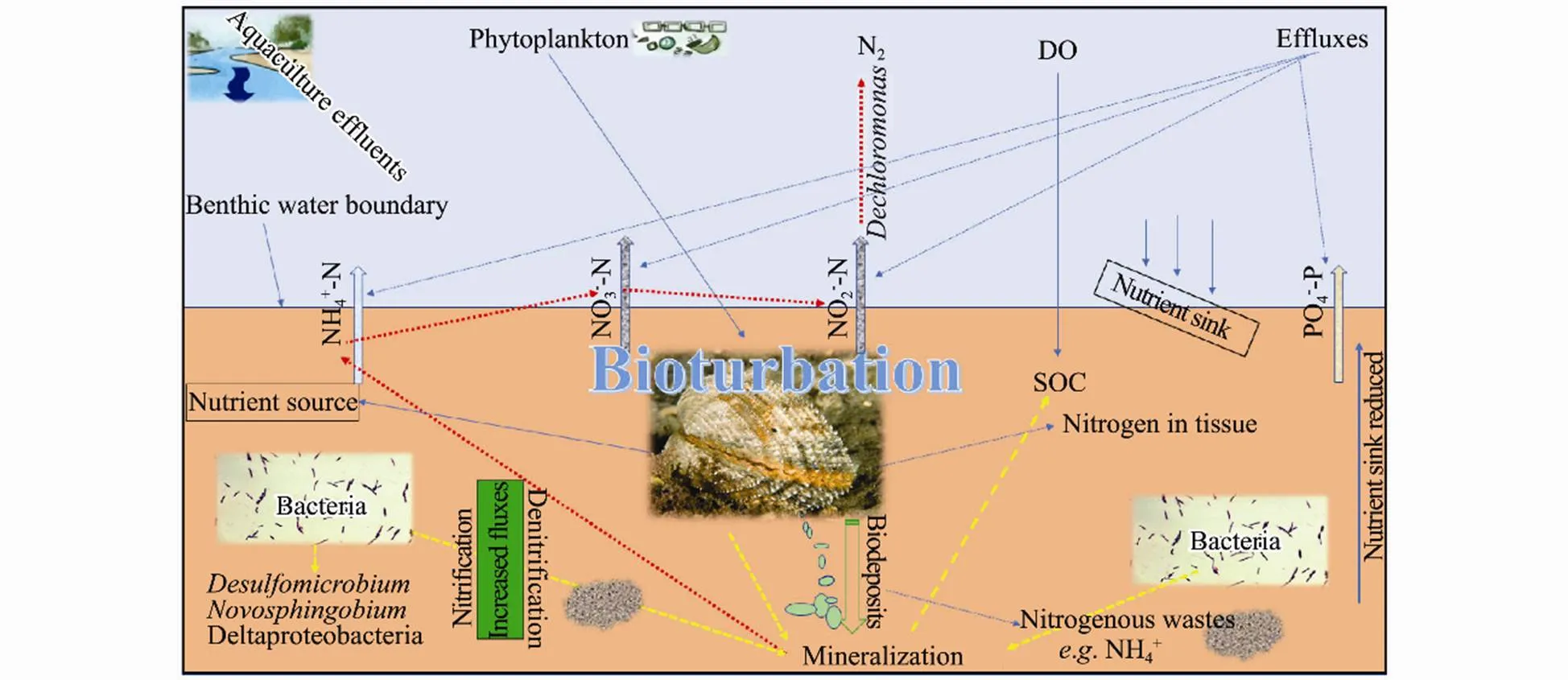
Fig.7 A hypothetical sketch of sediment-bacteria-macrofauna ecological processes involved in the nutrient dynamics in the bivalve bioturbated sediment ecosystem.
4.3 Relationship Between the Microbial Community and Nutrient Dynamics
Microorganisms contribute significantly to the balancing and cycling of marine biogenic elements (Azam and Malfatti, 2007; Dyksma., 2016). In our study, the first two ordination axes of CCA, and PCA plot explained 42.3%, and 22.9%, 73.2% and 22.9% of the total variation in the bacterial community structure (Fig.6a). The results of CCA showed, the environmental variables such as nutrient fluxes, TOC, and SOC parameters, were correlated with bacterial communities (genus level, relative abundance >3%). The correlations between bacterial communities and nutrient fluxes were high, indicating a strong relationship between the microbial community and environmental variables. Further, an association was revealed between several bacterial taxa, associated with nitrification and denitrification such as Proteobacteria, Nitrospirae, Firmicutes, Bacteroidetes, Pseudomonas, and the benthic nutrient fluxes (Figs.5, 6b). The presence of these taxa signifies that probably the dinitrogen nitrate reduction to ammonium activities in the clam sediments was promoted (Moncada., 2019). Potential sulfate and phosphate-reducing taxa such as Deltaproteobacteria and Desulfomicrobium) were promoted in this study (Fig.6a, b) Desulfomicrobium can contribute indirectly to inorganic nutrient release (.., PO43−-P) via organic matter mineralization (Sinkko., 2011; Ki., 2018). The CCA results indicated that TOC, OM, NH4+-N, and NO3−-N were among factors correlated with several types of sediment bacteria. The OM in the sediment is expected to decompose into phosphate (PO43−-P) by the sulfate-reducing bacteria (SRB), such as,,, and, which are found in both the upper and deeper sediment layers. In most samples, bacteria relating to key functions such as Desulfatibacillum, Burkholderiales, Desulfobacterium, Desulfomicrobium, and Desulfosalsimonas, were dominant, and positively expressed in the treatments (Figs.5, 6b).,,play a key role in decomposing organic phosphorus indirectly to phosphate via OM mineralization (Zhao., 2012; Ki., 2018). Therefore, the close correlation among the different microbial functional genes and strong correlations between PCA and CCA in the clam treatment may suggest a strong link between benthic nutrient cycling processes and the microbial community which is key aspect in bioremediation systems The diverse microbial community and increased nutrient dynamics in the clam treatment is due to the fact that ecosystem engineering processes by benthic macrofauna bioturbation can cause steep chemical gradients along burrows, resulting in accelerated mineralization, sulfide oxidation, sulfate reduction, and denitrification coupled nitrification activities (Meysman., 2006).has been reported to highly alter the microbial community structure and increased nutrients flux and cycling in benthic sediments (Shen., 2017; Lukwambe., 2019b; Nicholaus., 2019b). Other clams with similar activities such as(Liu., 2007),(Nicholaus and Zheng, 2014), and(Zhao., 2019) has been shown to contribute on benthic nutrient dynamics which impact bioremediation. In the current study, due to close similarity in morphology and burrowing behavior,has equally expressed more or less similar roles. It’s well-known that the transformation and cycling of biogeochemical nutrients such as nitrogen and phosphorus are driven by sediment microorganisms (Hou., 2013). Decomposition processes involvingandcan have a strong effect on ammonification pathways (Dauda., 2014). Fig.7 describes the existing relationship between sediment bacterial community degradation processes, sediment contents, andbioturbation activities. Since the presence ofpositively accelerated the microbial biomass turnover may suggest that mineralization processes of the nitrogen associated compounds (ammonium, nitrite, and nitrate) and other organic components were similarly accelerated thus prompting high flux rates, nutrient cycling, reduced organic wastes, and improved response of microbial activities to nutrient load.
5 Conclusions
The current study demonstrates the contribution of clamon the microbial community assemblages and nutrient dynamics in the sediments of the aquaculture bioremediation system. The clam treated area showed a higher diverse range of microbial community assembly. Specifically, the,,, andwere significantly expressed bacterial taxa among the treatments. Substantially theaccelerated the flux rate of NH4+-N, NO3−-N and PO43−-P and SOC. Moreover, organic contents (TP, TOC, TP and TN) especially from the surface cores in the clam treated sediments were significantly lower. Our findings suggest thatbioturbation activities predominantly increase microbial community diversity and nutrient dynamics, promotes organic content transformations, and improve bioremediation activities in the sediments by indirectly affecting the nitrification-denitri- fication activities, and modified sediment micro-habitats.
Acknowledgements
This study was supported by the Zhejiang Public Welfare Technology Research Program of China (ZPWTP) (No. LGN18C190008),the Zhejiang Provincial Natural Science Foundation of China (No. LQ20C190003), the Ningbo Municipal Natural Science Foundation (Nos. 2019A610 421, 2019A610443), and the K. C. Wong Magna Fund in Ningbo University.
Aller, R. C., 2001. Transport and reactions in the bioirrigated zone. In:. Boudreau, B., and Jørgensen, B. B., eds., Oxford University Press, New York, 269-301.
Anderson, M. J., 2001. A new method for non-parametric multivariate analysis of variance., 26: 32-46.
Azam, F., and Malfatti, F., 2007. Microbial structuring of marine ecosystems., 5: 782-791.
Bender, J., and Phillips, P., 2004. Microbial mats for multiple applications in aquaculture and bioremediation., 94: 229-238.
Bertics, V. J., and Ziebis, W., 2009. Biodiversity of benthic microbial communities in bioturbated coastal sediments is controlled by geochemical microniches., 3: 1269-1285.
Booth, J. M., Fusi, M., Marasco, R., Mbobo, T., and Daffonchio, D., 2019. Fiddler crab bioturbation determines consistent changes in bacterial communities across contrasting environmental conditions., 9 (1): 3749.
Brito, L. O., Cardoso Junior, L. de O., Lavander, H. D., Abreu, J. L., de Severi, W., and Gálvez, A. O., 2018. Bioremediation of shrimp biofloc wastewater using clam, seaweed, and fish., 34 (10): 901-913.
Caporaso, J. G., Bittinger, K., Bushman, F. D., DeSantis, T. Z., Andersen, G. L., and Knight, R., 2010. PyNAST: A flexible tool for aligning sequences to a template alignment., 26: 266-267.
Chen, X., Andersen, T. J., Morono, Y., Inagaki, F., Jørgensen, B. B., and Lever, M. A., 2017. Bioturbation as a key driver behind the dominance of bacteria over archaea in near-surface sediment., 7 (1): 2400.
Dale, A. W., and Prego, R., 2002. Physico-biogeochemical controls on the benthic-pelagic coupling of nutrient fluxes and recycling in a coastal upwelling system., 235: 15-28.
Dauda, A. B., and Akinwole, A. O., 2014. Interrelationships among water quality parameters in recirculating aquaculture system., 8: 20-25.
DeSantis, T. Z., Hugenholtz, P., Keller, K., Brodie, E. L., Larsen, N., Piceno, Y. M., Phan, R., and Andersen, G. L., 2006. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes., 34: 394-399.
Diaz, R. J., 2001. Overview of hypoxia around the world., 30: 275-281.
Dyksma, S., Bischof, K., Fuchs, B. M., Hoffmann, K., Meier, D., Meyerdierks, A., Pjevac, P., Probandt, D., Richter, M., and Stepanauskas, R., 2016. Ubiquitousdominate dark carbon fixation in coastal sediments., 10: 1939-1953.
Edgar, R. C., 2010. Search and clustering orders of magnitude faster than BLAST., 26 (19): 2460-2461.
Feng, S., Xie, S., Zhang, X., Yang, Z., Ding, W., Liao, X., Liu, Y., and Chen, C., 2012. Ammonium removal pathways and microbial community in GAC-sand dual media filter in drinking water treatment., 24: 1587-1593.
Hallett, L. M., Jones, S. K., MacDonald, A. A. M., Jones, M. B., Flynn, D. F. B., Ripplinger, J., Slaughter, P., Gries, C., and Collins, S. L., 2016. An r package of community dynamics metrics., 7: 1146-1151.
Hou, J., Song, C., Cao, X., and Zhou, Y., 2013. Shifts between ammonia-oxidizing bacteria and archaea in relation to nitrification potential across trophic gradients in two large Chinese lakes (Lake Taihu and Lake Chaohu)., 47: 2285-2296.
Ieno, E. N., Solan, M., Batty, P., and Pierce, G. J., 2006. How biodiversity affects ecosystem functioning: Roles of infaunal species richness, identity and density in the marine benthos., 311: 263-271.
Karakassis, I., Tsapakis, M., Hatziyanni, E., Papadopoulou, K. N., and Plaiti, W., 2000. Impact of cage farming of fish on the seabed in three Mediterranean coastal areas., 57: 1462-1471.
Kautsky, N., and Evans, S., 1987. Role of biodeposition byin the circulation of matter and nutrients in a Baltic coastal ecosystem., 38: 201-212.
Ki, B. M., Huh, I. A., Choi, J. H., and Cho, K. S., 2018. Relationship of nutrient dynamics and bacterial community structure at the water-sediment interface using a benthic chamber experiment., 53 (5): 482-491.
Kristensen, E., and Kostka, J. E., 2005. Macrofaunal burrows and irrigation in marine sediment: Microbiological and biogeochemical interactions. In:. Kristensen, E.,., eds., American Geophysical Union, Washington, D. C., 125-157.
Learman, D. R., Henson, M. W., Thrash, J. C., Temperton, B., Brannock, P. M., Santos, S. R., Mahon, A. R., and Halanych, K. M., 2016. Biogeochemical and microbial variation across 5500km of Antarctic surface sediment implicates organic matter as a driver of benthic community structure., 7: 284.
Liu, J., Chen, Z., Xu, S., and Zheng, X., 2007. Experimental research on the impact ofon DIN exchange at a tidal flat sediment-water interface., 25 (4): 434-443.
Lukwambe, B., Nicholaus, R., Yang, W., and Zheng, Z., 2019a. Blood clams()bioturbation alter succession of bacterioplankton community and nutrients removal performance in an aquaculture wastewater bioremediation system.,734520.
Lukwambe, B., Nicholaus, R., Zhao, L., Yang W., Zhu, J., and Zheng, Z., 2020. Microbial community and interspecies interaction during grazing of ark shell bivalve () in a full-scale bioremediation system of mariculture effluents., 158: 104956.
Lukwambe, B., Yang, W., Zheng, Y., Nicholaus, R., Zhu, J., and Zheng, Z., 2018. Bioturbation by the razor clam () on the microbial community and enzymatic activities in the sediment of an ecological aquaculture wastewater treatment system., 643: 1098-1107.
Lukwambe, B., Zhao, L., Nicholaus, R., Yang, W., Zhu, J., and Zheng, Z., 2019b. Bacterioplankton community in response to biological filters (clam, biofilm, and macrophytes) in an integrated aquaculture wastewater bioremediation system., 254: 113035.
Magoč, T., and Salzberg, S. L., 2011. FLASH: Fast length adjustment of short reads to improve genome assemblies., 27: 2957-2963.
Mermillod-Blondin, F., Rosenberg, R., François-Carcaillet, F., Norling, K., and Mauclaire, L., 2004. Influence of bioturbation by three benthic infaunal species on microbial communities and biogeochemical processes in marine sediment., 36: 271-284.
Meysman, F. J. R., Middelburg, J. J., and Heip, C. H. R., 2006. Bioturbation: A fresh look at Darwin’s last idea., 21: 688-695.
Moncada, C., Hassenrück, C., Gärdes, A., and Conaco, C., 2019. Microbial community composition of sediments influenced by intensive mariculture activity., 19 (12): 1-12.
Moriarty, D. J. W., 1997. The role of microorganisms in aquaculture ponds., 151 (1-4): 333-349.
Mudrich, A., Azcue, J. M., and Mudroch, P., 1996.. CRC Press Inc., Boca Raton, Florida, USA, 1-12.
Navarrete-Mier, F., Sanz-Lázaro, C., and Marín, A., 2010. Does bivalve mollusc polyculture reduce marine finfish farming environmental impact?, 306 (1-4): 101-107.
Nicholaus, R., and Zheng, Z. M., 2014. The effects of bioturbation by the Venus clamon the fluxes of nutrients across the sediment-water interface in aquaculture ponds., 22 (2): 913-924.
Nicholaus, R., Lukwambe, B., Lai, H., Yang, W., and Zheng, Z., 2019a. Nutrients cycling in ecological aquaculture wastewater treatment systems: Vertical distribution of benthic phosphorus fractions due to bioturbation activity by, 11: 469-480.
Nicholaus, R., Lukwambe, B., Zhao, L., Yang, W., Zhu, J., and Zheng, Z., 2019b. Bioturbation of blood clamon benthic nutrient fluxes and microbial community in an aquaculture wastewater treatment system., 142:73-82.
Papaspyrou, S., Gregersen, T., Kristensen, E., Christensen, B., and Cox, R., 2006. Microbial reaction rates and bacterial communities in sediment surrounding burrows of two nereidid polychaetes (and)., 148: 541-550.
R Core Team, 2017. R: A Language and Environment for Statistical Computing. https://www.R-project.org/.
Satoh, H., Nakamura, Y., and Okabe, S., 2007. Influences of in- faunal burrows on the community structure and activity of ammonia-oxidizing bacteria in intertidal sediments., 73: 1341-1348.
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., and Huttenhower, C., 2011. Metagenomic biomarker discovery and explanation., 12: R60.
Shen, H., Jiang, G., Wan, X., Li, H., Qiao, Y., Thrush, S., and He, P., 2017. Response of the microbial community to bioturbation by benthic macrofauna on intertidal flats., 488: 44-51.
Sinkko, H., Lukkari, K., Jama, A. S., Sihvonen, L. M., and Si- vonen, K., 2011. Phosphorus chemistry and bacterial community composition interact in brackish sediments receiving agricultural discharges., 6: e21555.
Solan, M., Batty, P., Bulling, M. T., and Godbold, J. A., 2008. How biodiversity affects ecosystem processes: Implications for ecological revolutions and benthic ecosystem function., 2: 289-301.
Stauffert, M., Duran, R., Gassie, C., and Cravo-Laureau, C., 2014. Response of archaeal communities to oil spill in bioturbated mudflat sediments., 67: 108-119.
Volkenborn, N., Polerecky, L., Wethey, D. S., DeWitt, T. H., and Woodin, S. A., 2012. Hydraulic activities by ghost shrimpinduce oxic-anoxic oscillations in sediments., 455: 141-156.
Yang, J., Gou, Y., Fang, F., Guo, J., Lu, L., Zhou, Y., and Ma, H., 2018. Potential of wastewater treatment using a concentrated and suspended algal-bacterial consortium in a photo mem- brane bioreactor., 335: 154- 160.
Zhang, L., Shen, Q. S., Hu, H. Y., Shao, S. G., and Fan, C. X., 2011. Impacts ofon oxygen uptake and nutrient fluxes across the sediment-water Interface., 220: 399-411.
Zhang, S., Fang, X., Zhang, J., Fang, Y., Hu, Z., Lizhen, W., and Daisuke, K., 2020. The effect of bioturbation activity of the ark clamon the fluxes of nutrient exchange at the sediment-water interface., 19: 232-240.
Zhao, G., Du, J., Jia, Y., Lv, Y., Han, G., and Tian, X., 2012. The importance of bacteria in promoting algal growth in eutrophic lakes with limited available phosphorus., 42: 107-111.
Zhao, L., Zheng, Y., Nicholaus, R., Lukwambe, B., Zhu, J., Yang, W., and Zheng, Z., 2019. Bioturbation by the razor clamaffects benthic nutrient fluxes in aquaculture wastewater treatment ecosystems., 11: 87-96.
Zheng, Z. M., Dong, S. L., and Bai, P. F., 2009. Study on nutrient fluxes across sediment-water interface in different sea cucumber experimental enclosures., 39: 209-214 (in Chinese with English abstract).
. E-mail:zhengzhongming@nbu.edu.cn
January 21, 2020;
May 10, 2020;
September 24, 2020
(Edited by Ji Dechun)
杂志排行
Journal of Ocean University of China的其它文章
- Numerical Simulation and Risk Analysis of Coastal Inundation in Land Reclamation Areas: A Case Study of the Pearl River Estuary
- Variation of Yellow River Runoff and Its Influence on Salinity in Laizhou Bay
- Cold Water in the Lee of the Batanes Islands in the Luzon Strait
- Preliminary Design of a Submerged Support Structure for Floating Wind Turbines
- Inversion of Oceanic Parameters Represented by CTD Utilizing Seismic Multi-Attributes Based on Convolutional Neural Network
- Trace-Norm Regularized Multi-Task Learning for Sea State Bias Estimation
