Gut microbiota and non-alcoholic fatty liver disease
2015-12-24
Athens, Greece
Gut microbiota and non-alcoholic fatty liver disease
Paraskevas Gkolfakis, George Dimitriadis and Konstantinos Triantafyllou
Athens, Greece
A "two hits" hypothesis about NAFLD pathogenesis has been proposed.[14]The first hit is increased triglycerides accumulation in the liver, whereas a second one (e.g. oxidative stress) induces liver parenchyma inflammation leading to NASH. Recently, investigators have proposed the "multiple parallel" hits hypothesis:[15,16]inflammation may either precede or follow simple steatosis with multiple factors, namely lipotoxicity, increased oxidative stress, mitochondrial dysfunction and iron overload acting in parallel to promote NASH.[16]Moreover, a genetic predisposition to the disease is possible since a mutation in the patatin-like phospholipase domain-containing 3 gene has been recognized to strongly predict liver fat accumulation and disease progression.[17]
The aim of this review is to highlight key issues on gut microbiota-host crosstalk regarding the pathogenesis of NAFLD and NASH. We present herein the yield of a PubMed search from 1995 to 2014 using the key words "non-alcoholic fatty liver disease", "non-alcoholic steatohepatitis", "fatty liver", "gut microbiota" and "microbiome".
Gut microbiome
Gut microbiota is a group of commensal microorganisms that live synergistically with the host. They process complex, otherwise indigestible, polysaccharides to short-chain fatty acids, thus providing extra energy for the host. They also participate in the synthesis of vitamins (e.g. vitamin K) and in the development and maintenance of the immunity at the intestinal lumen level.[18,19]
The adult type gut microbiota is made up from bacteria, viruses, protozoa, archaea, eukaryotes, yeasts and parasites. It counts more than 1014cells,[20]more than 100 different bacterial species;[18]their genome counts up to 300 000 genes,[21]100 times the number of the human genome. Among them, bacteria predominate with the Gram-positive short-chain fatty acids-producing firmicutes and the Gram-negative hydrogen-producing bacteroidetes being the main phyla, followed by proteobacteria, actinobacteria, bifidobacteria, etc.[22,23]Based on the abundant genera, two basic enterotypes are recognized:[24]Enterotype 1, wherebacteroides spp.dominate and enterotype 2, with abundance ofprevotella spp.. The existence of a third enterotype--enterotype H[25]--with abundance of bothbacteroides spp.andprevotella spp.has also been proposed.
Evaluating gut microbiota in NAFLD
In order to reveal microbiota composition, culturedependent and culture-independent techniques have been implicated.[26]Traditional culture allows detection and semi-quantification of many bacterial groups. Nevertheless, since a large amount of gut bacteria requires special conditions (e.g. anaerobic environment) to grow, a loss of 80% of the detectable bacteria is anticipated.[22]Trying to overcome this problem, culture-independent techniques have been developed. Apart from the widely used quantitative real-time polymerase chain reaction (qRT-PCR), additional techniques based on the diversity in the sequence of the bacterial 16S ribosomal RNA (16S rRNA) gene are now utilized: either the entire or conserved regions of the 16S rRNA gene are amplified, the results are compared with the sequence-containing libraries, thus accurate bacterial species identification or the partial 16S rRNA sequencing (pyrosequencing) provides information about the number, nature and abundance of the diverse bacterial species[27]without need of anex vivobacterial culture or DNA cloning.
Furthermore, the "-omics" studies are reliable methods to specify the functional species that interact with the host. Accordingly, the entire bacterial genome (metagenomics), the expressed mRNA (metatranscriptomics), the obtained proteins from the investigated microenvironment (metaproteomics), and the produced metabolites (metabolomics) are used to distinguish different phenotypes as well as, potential host-microbiome interactions.
Table 1 summarizes data from human studies investigating stool microbiota in NAFLD. Zhu et al[25]studied a pediatric population of 22 biopsy-proven NASH subjects, 25 obese children and 16 healthy controls using 16S rRNA sequencing in stool samples. Sequencing revealed a significant increase in bacteroidetes with decreased levels of firmicutes among obese and NASH individuals. Actinobacteria were significantly decreased in NASH patients, whereas proteobacteria counts showed a gradual increase from healthy to obese and NASH patients. Authors also provided evidence of increased ethanol levels in NASH children, postulating a possible role of abundant ethanol-producing bacteria (such asEscherichia) in NASH progression.
The increased bacteroidetes/firmicutes ratio detected by Zhu et al[25]was not confirmed by Raman et al,[28]who studied 30 obese adult patients with clinical, biochemical and radiological suspicion of NAFLD and 30 healthy controls using multi-tag pyrosequencing in a single stool sample per patient. Although differences in family and genera level were detected, the authors could not demonstrate a significant difference in the phylum level between the two groups. Similarly, using qRT-PCR in stool samples of 33 patients with biopsy-proven NAFLD (22 with NASH) and 17 healthy controls, Mouzaki et al[29]revealed significantly lower counts of Bacteroides in NASH patients than patients with simple steatosis and healthy controls. Firmicutes, proteobacteria and actino-bacteria counts were not different significantly among the groups, whereas archaea were detectable only in 9/50 subjects (2 with simple steatosis, 2 with NASH and 5 healthy controls).
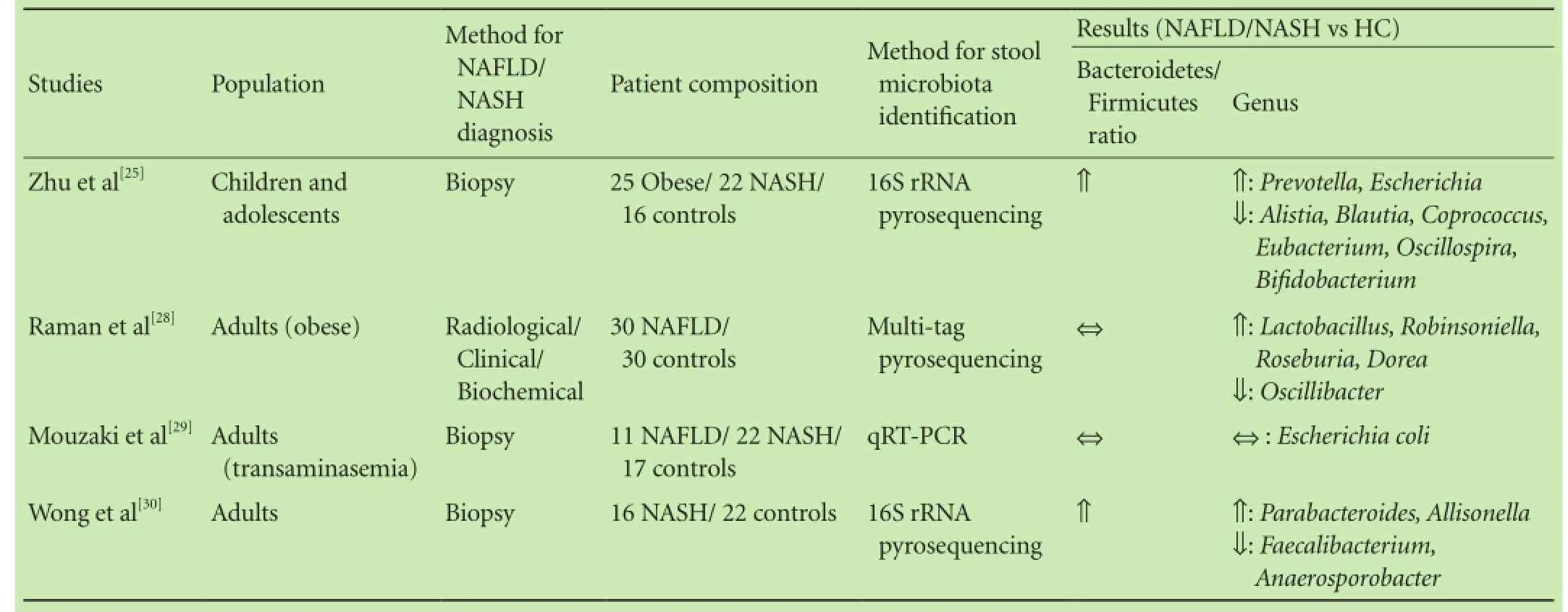
Table 1. Studies on stool microbiota in NAFLD
Trying to clarify the impact of pro- and prebiotics on gut microbiota composition, Wong et al[30]evaluated biopsy-proven NASH patients and controls. Participants provided stool samples for microbial counts measurements using 16S rRNA sequencing. Bacteroidetes were the dominant phyla in both groups (NASH and healthy volunteers), followed by firmicutes, which showed a notable presence in healthy controls.
Finally, the effect of choline-deficient diet, a mechanism implicated in NAFLD pathogenesis, was studied by Spencer et al.[31]Stool samples were collected in predefined time points and 16S rRNA pyrosequencing was used for microbiota specification. The authors did not find any common change in the taxons' abundance in the different time points of the study. However, the dramatically fall to zero inGammaproteobacteria(phylum Proteobacteria) counts and the strong correlation of baseline levels ofGammaproteobacteriato liver fat enrichment during the choline-deficient diet postulated thatGammaproteobacterialevels may predict the possibility of an individual to develop fatty liver when exposed to choline-deficient diet.
Taken together, the above studies provide different evidence regarding NAFLD/NASH and gut microbiota composition. Bacteroidetes/firmicutes ratio is increased in NASH children, while no change is revealed in NAFLD/NASH adults compared to healthy controls. This discrepancy at the phylum level raises questions about a potential role of microbiota in NAFLD/NASH development. When focusing at the genera, a wide diversity regarding flora composition can be detected. This finding can be attributed to different methods used or even different studied populations; however, it is unlikely to extract safe conclusions regarding a potential causative role of special phyla and genera in NAFLD/NASH development.
Potential mechanisms that implicate gut microbiota in the pathogenesis of NAFLD
Accumulating data from preclinical and clinical studies suggest a critical role of gut microbiota in NAFLD pathogenesis, mainly through predisposition to obesity, metabolism alterations (insulin resistance) and promoted liver inflammation. Moreover, many bacterial byproducts, such as ethanol, might demonstrate hepatotoxicity via stimulation of Kupffer cells to produce and exude nitric acid and cytokines[32](Fig.).
Obesity
More than a decade ago, investigators showed that germ-free mice gained 42% less weight in comparison with animals that hosted gut microbiota, despite the fact that they consumed more calories.[33]Moreover, when cecal microbiota was transplanted from normal to germfree mice, the animals gained 57% more weight without any change in consumed calories. The same group emphasized their results by showing that germ-free mice were unable to gain weight while maintaining a high-calorie diet.[34]
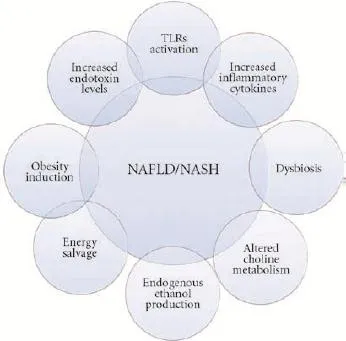
Fig. Potential mechanisms through which gut microbiota contribute to NAFLD/NASH pathogenesis. Overlapping external circles indicate common pathogenic mechanisms. NAFLD: non-alcoholic fatty liver disease; NASH: non-alcoholic steatohepatitis; TLRs: Toll-like receptors.
Turnbaugh et al[35]transplanted gut microbiota from obese ob/ob mice and lean ob/+ or +/+ mice to lean germ-free mice, respectively. Animals that received microbiota from obese donors showed a higher fat gain than those that received microbiota from lean donors. Obesity-associated gut microbiota was able to extract more energy from the diet via digesting otherwise indigestible polysaccharides into short-chain fatty acids. Further supporting the aforementioned hypothesis, the total fecal short-chain fatty acids concentration from 33 obese human individuals was 20% higher than that from 30 lean volunteers.[36]
A potential role of specific gut microbiota in obesity and subsequent NAFLD development has been proposed. Obese mice hosted 50% less bacteroidetes and more firmicutes as compared with lean controls, whereas archaea counts were significantly increased in the obese animals.[37]
The decreased bacteroidetes/firmicutes ratio was also detected in obese human studies. Overweighed and obese children at the age of 7 had less bacteroidetes and moreS. aureusin stools collected at the age of 6 and 12 months of age.[38]A study of 12 obese individuals revealed more bacteroidetes and less firmicutes, but this ratio was reversed when they followed a fat or carbohydrate restricted diet for 1 year.[39]On the contrary, firmicutes and archaea showed lower counts in obese patients after bariatric surgery.[40]Enterotype 1 has been associated with the long-term consumption of animal proteins and saturated fat, whereas diet based on carbohydrates predisposes to Enterotype 2.[41]These data underline that the role of gut microbiota diversity pends to be further clarified in obesity.
In contrast, a recently described bacterium[42]Akkermansia municiphilahas been associated with non-obese phenotype. Pregnant women who gained excess weight during pregnancy[43]as well as obese and overweight preschool children[44]had low fecal concentrations ofA. municiphila. Similarly,A. municiphilacounts were markedly increased in obese mice that underwent Roux-Y gastric bypass surgery.[45]
Experimental models provided evidence for a modulating role ofA. municiphilain weight gain, type 2 diabetes mellitus and eventually NAFLD. "Treatment" of mice under high-fat-diet withA. municiphilaled to induction of Tregs cells in the visceral adipose tissue. This resulted in the attenuation of adipose tissue inflammation and increased glucose tolerance, effects that resemble the antidiabetic function of metformin.[46]Similarly, Everard et al[47]showed thatA. municiphilaadministration in type 2 diabetes mellitus mice led to a gradual reversion of insulin resistance, adipose tissue inflammation, fat gain and endotoxemia.
Ethanol
Endogenous ethanol is a metabolite of many gut microbiota species. After its absorption, it reaches the liver via the portal vein.[48]Alcohol dehydrogenase catalyzes its oxidation in the liver, leading to acetate and acetaldehyde formation.[49]The first is a substrate for fatty acids synthesis, and the second produces reactive oxygen species. Therefore, ethanol metabolism induces triglycerides accumulation in the liver[50]and hepatic oxidative stress, thus fulfilling both steps of the "two hits" hypothesis.
Increased ethanol levels have been detected in obese patients[51]as well as, in non-alcohol consuming children with NASH,[25]indicating its causative role in NAFLD/ NASH development. Moreover, significantly increased expression of the ethanol-metabolizing enzymes alcohol dehydrogenase, catalase and aldehyde dehydrogenase has been detected in NASH livers.[52]Ethanol may also promote NAFLD by increasing gut mucosal permeability[53]that induces endotoxemia:[54]five days ciprofloxacin administration in NASH patients with ethanol detected in their blood resulted in immeasurable levels of ethanol,[55]pointing to a close relationship of endogenous ethanol production and gut microbiota.
Lipopolysaccharide-endotoxemia-Toll-like receptors
Endotoxin is part of the Gram-negative bacterial cellmembrane. Lipopolysaccharide (LPS), the active component of endotoxin, binds to the LPS-binding protein and its CD14 receptor[56]to form a complex that interacts with Toll-like receptors (TLRs) and activates inflammatory cascade.[57]
Genetically obese mice develop steatohepatitis after infusion of low doses of LPS.[58]Furthermore, LPS injected in NAFLD mice further promotes liver injury by enhancement of produced proinflammatory cytokines,[59]whereas high-fat-diet induces increased plasma circulating LPS.[60]Diet-induced NAFLD in rodents is also associated with increased levels of LPS.[60,61]In human studies, subjects with NAFLD experience significantly higher levels of circulating endotoxin,[62,63]with marked increases in early fibrosis, when compared with healthy controls.
TLRs activation leads to the translocation of NF-κB in the nucleus and induction of proinflammatory genes transcription, such as TNF-α, IL-1β, IL-6 and IL-12.[64]IL-1β augments triglycerides accumulation in the hepatocytes by enhancement of diacyloglycerol transferase that converts diglycerides into triglycerides.[65]Moreover, TNF-α inhibits insulin receptors as well as insulin-receptors-substrate-1, leading to increased levels of circulating insulin and therefore to insulin resistance.[62]Consequently, influx of fatty acids derived from adipose tissue is facilitated.[66]
Recently, inflammasomes deficiencies have been implicated in NAFLD development. Inflammasomes are cytoplasmic multiprotein complexes that regulate the cleavage of pro-IL-1β and pro-IL-18 into the respective cytokines, through activation of caspase-1.[48]Genetically deficient for components of the inflammasome (NLRP3 and NLRP6) mice had higher levels of LPS and bacterial DNA in the portal circulation. The former binds to TLR-4 and the latter to TLR-9, leading to increased expression of TNF-α in the liver, thus promoting liver inflammation and steatosis.[67]
To summarize, NAFLD and NASH are characterized by increased levels of endotoxin. Endotoxin, one among other components of the second hit, in the "two hits" hypothesis triggers a cascade to increased levels of proinflammatory cytokines, insulin resistance and triglycerides production, thus facilitating NAFLD development and progression. Moreover, TLRs- and inflammasomedeficient mice showed either NAFLD or steatosis worsening, strengthening the role of the endotoxin-path in NAFLD pathogenesis. liver.[68]Gut microbiota are implicated in choline metabolism by producing enzymes that catalyze choline into methylamines, which potentially induce inflammation when absorbed by the liver.[69]Metabolomics analysis in 129S6 mouse strain animals with high-fat-diet induced steatosis revealed a significant decrease in circulating phosphatidylcholine and increased urine excretion of its metabolites, thus supporting the presence of a gut microbiota phenotype that induces choline deficiencymediated liver injury.[70]Apart from that, choline deficiency contributes to triglycerides accumulation in the liver and decreased liver secretion of very-low-density lipoprotein,[48]whereas choline deficient diet resulted in liver steatosis that reversed after choline substitution in animals.[71]Acknowledging the fact that the data derive from a deficient disease and therefore the model might not be suitable enough to explain NAFLD development, decreased choline levels and increased levels of toxic choline metabolites might represent the gut microbiota choline-deficiency-mediated mechanism in NAFLD development.
Dysbiosis
Investigators compared metabolic and histologic changes in mice receiving microbiota from donors with different metabolic phenotypes.[72]Mice receiving intestinal microbiota from donors with hyperglycemia and increased levels of proinflamamtory cytokines developed hyperglycemia, increased levels of insulin and macrovesicular steatosis, whereas mice receiving microbiota from normoglycemic donors did not show similar metabolic and histologic changes. Dysbiosis, namely increased counts of firmicutes in the former and increased population ofBacteroides vugatusin the latter receivers, was detected by means of 16S rRNA pyrosequencing techniques.
Choline
Choline, a phospholipid component of cell membrane, plays a crucial role in lipid transport from the
Small intestinal bacterial overgrowth - intestinal permeability
Small intestinal bacterial overgrowth (SIBO) is the presence of increased number of colonic type anaerobic and/ or aerobic bacteria in the small intestine.[23]In combination with intestinal permeability, SIBO has been proposed as a pathogenetic mechanism of NAFLD/NASH development. Ectopic microbiota in SIBO may cause loose junctions in the small intestinal epithelium facilitating the flow of bacterial byproducts in the portal vein system and their absorption from the liver, where they manifest their toxic activities. There is no gold standard technique to diagnose SIBO yet, since even aspirate culture suffers from certain methodological caveats. Breathtesting is the current diagnostic test for SIBO, whereas culture independent techniques like qRT-PCR and deep sequencing have also been used in this setting.[23]
The correlation of NAFLD and SIBO has been investigated in experimental[73]and human studies[53,55,74-79](Table 2). Increased prevalence of SIBO in NAFLD/NASH humans[74,78]and in a rat NASH model[73]has been indirectly documented by measuring the orocecal transit time.
In obese bariatric surgery patients, SIBO detected using hydrogen breath testing has been revealed as an independent factor for severe steatosis in liver biopsy.[76]Wigg et al[79]detected SIBO in 50% of NASH patients, in whom higher levels of TNF-α were also identified and lactulose breath test detected SIBO in 78% of NASH patients in association with TLR-4 expression and plasma levels of IL-8.[77]In contrast to the aforementioned data, among 20 NAFLD patients SIBO was identified in only 3 using glucose breath test.[53]Using the lactulose/mannitol test that assesses intestinal permeability, a higher index of intestinal permeability as calculated by the lactulose/mannitol urine excretion ratio was demonstrated in NAFLD patients.[53]Similarly, Miele et al[75]detected SIBO in 60% of 35 biopsy-proven NAFLD patients and a significant correlation between the severity of steatosis and increased intestinal permeability was evident. Moreover, patients with NAFLD and increased intestinal permeability had a 2-fold higher prevalence of SIBO in comparison with patients with normal intestinal permeability. Authors could not support a causative role of SIBO in the progression of NAFLD, since they did not reveal correlation between increased intestinal permeability and/or SIBO with steatohepatitis.
The role of archaea
Methanogenic archaea scavenge hydrogen (H) and ammonia (NH4) to produce methane (CH4). Hydrogen utilization optimizes microenvironmental intestinal lumen conditions that facilitate short-chain fatty acids production and salvage energy for the host by gut flora.[80]
Methanobrevibacter smithii(M. smithii) is the predominant methanogenic archaeon detected in 70% of healthy individuals.[81]A causative role ofM. smithiifor obesity development has been proposed in a mouse model. Colonization either withM. smithiiandBacteroides thetaiotaomicronorB. thetaiotaomicronalone led to increased weight gain only in subjects colonized withM. smithii.[82]
Methane detection in breath test is associated with increased levels ofM. smithiiin stools,[83]setting this cheap and non-invasive method as a guide for further studies. Indeed, when methane-positive obese subjects were compared with obese methane-negative controls, they showed a 6.7 kg/m2body-mass-index (BMI) increase.[84]Moreover, when methane and hydrogen weremeasured in 792 subjects using lactulose breath test, methane-positive and hydrogen-positive (M+/H+) individuals showed increased BMI and increased percent body fat compared with the other groups (M-/H-, M+/H-, M-/H+).[85]It was therefore proposed that hydrogen derived from increased microbiota fermentation acts as a tank providing fuel to hydrogen-required metabolism of methanogens. In order to strengthen the hypothesis thatM. smithiiinfluences obesity development, Mathur et al[86]correlated the location and extent ofM. smithiicolonization with weight gain in rats. Using qRT-PCR, investigators showed thatM. smithiicounts were higher in the small intestine (highest in the ileum), whereas total bacterial numbers were the lowest in the small intestine and the highest in the left colon and cecum. Of interest, mice fed high-fat-diet had a significantly increasedM. smithiiconcentration in the duodenum, ileum and cecum, whereas no difference was found in the total bacterial counts. Furthermore, animals with all five intestinal parts colonized withM. smithiihad higher body weight than those with limited colonization.

Table 2. Small intestinal bacterial overgrowth in NAFLD
In conclusion, these studies provide evidence for the first time that methanogenic archaea counts, especiallyM. smithiithat colonize not only the colon but also the small intestine correlate with diet-induced weight gain. placebo,[91]Lactobacillus GGvs placebo,[92]Bifidobacterium longumwith fructooligosaccharides vs placebo,[93]lepicol probiotic formula vs nothing[94]) and a variety of administration periods, a meta-analysis[95]provided evidence that probiotics might have a positive effect on biochemical markers in patients with NAFLD and NASH. More specifically, the use of probiotics was associated with a significant reduction of ALT, AST, cholesterol, TNF-α and homeostasis model assessment of insulin resistance. The amelioration of these parameters using probiotics creates promises for further scientific evaluation.
Probiotics as a novel therapeutic approach
Probiotics are "live microorganisms that, when administered in adequate amounts confer a health benefit on the host".[87]Studies in mice and rodents revealed a potential protective role of two Lactobacillus strains:Lactobacillus rhamnosus GG[88]andLactobacillus casei Shirota,[89]in diet-induced NAFLD/NASH by demonstrating biochemical and histological attenuation of liver steatosis. Moreover, probiotics altered gut microbiota and prevented NAFLD progression in animals.[90]In humans, few lowquality randomized clinical trials investigated the potential therapeutic role for probiotics in patients with fatty liver. Wong et al[30]randomized 20 biopsy-proven NASH patients to receive either a combination of prebiotic and probiotic formula (containingLactobacillus spp.andBifidobacterium spp.) or prebiotic alone. Participants provided stool samples for microbial counts measurements using 16S rRNA sequencing, at baseline and at month six. Measurement of intrahepatic triglyceride content by spectroscopy was used to assess liver steatosis. Decreased counts of firmicutes and increased counts of Bacteroidetes were observed at 6 months in NASH patients experiencing lower levels of intrahepatic triglycerides content.
Moreover, despite different probiotics mixtures (Lactobacillus bulgaricusandStreptococcus thermophilusvs
Perspectives
Gut microbiota is currently studied to elucidate its role in the pathophysiology of NAFLD. Excessive energy harvest, ethanol production, LPS-TLRs interaction and choline metabolism alterations are the main principal mechanisms implicated so far. Progress in molecular DNA-based techniques allows a more detailed characterization of this microbiota. While the gold standard for bacterial enumeration remains the qRT-PCR,[23]one of its disadvantages is that unknown species cannot be identified. In these terms, development of metagenomic techniques (16S rRNA, multi-tag sequencing) opens a promising window to investigate the microbiome. The aforementioned studies aimed to reveal a potential correlation between gut microbiota composition and NAFLD development or progression to NASH. Despite interesting findings regarding abundant phyla and genera, the results show wide discordance and heterogeneity. Factors that could explain these differences include selection criteria for the study populations, variable methodologies in defining NAFLD, unadjusted diet-induced manipulation of microbiota, medications intake that may alter gut microbiota, such as proton pump inhibitors,[96]as well as the DNA-based techniques' inability to distinguish whether the genetic material comes from alive and active or dead and inactive microorganisms.
Trying to temper these difficulties, RNA-, mRNA-and protein-based techniques (metagenomics, metatranscriptomics and metaproteomics) have been developed. Despite their limited availability, a future combination and standardization of these techniques might lead to deeper comprehension of the microbiota role in NAFLD.
Another issue that should be highlighted is that in the vast majority of the studies, only stool samples were examined. Isolate stool sample analysis may underestimate or even cloak the contribution of small intestinal flora and small bacterial overgrowth in NAFLD pathophysiology.[97]Advanced, culture-independent techniques applied directly on duodenal and small intestinal contentand biofilm may uncover a potential region-dependent interaction between liver steatosis and gut microbiota. Moreover, in both experimental models and in humans, the role of archaea, yeasts and viruses in NAFLD pathogenesis remains largely uninvestigated.
Finally, there is limited evidence for microbiota manipulation with probiotics as prevention or therapy of NAFLD.
Acknowledgment:We acknowledge Dr. AD Sioulas' critical evaluation of the manuscript.
Contributors:GP searched the literature and drafted the manuscript. DG reviewed the draft. TK conceived the idea and reviewed the draft. All authors approved the final version. TK is the guarantor.
Funding:None.
Ethical approval:Not needed.
Competing interest:No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
1 Bugianesi E, McCullough AJ, Marchesini G. Insulin resistance: a metabolic pathway to chronic liver disease. Hepatology 2005; 42:987-1000.
2 Bhala N, Jouness RI, Bugianesi E. Epidemiology and natural history of patients with NAFLD. Curr Pharm Des 2013;19: 5169-5176.
3 Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology 2012;55:2005-2023.
4 Duseja A, Chawla YK. Obesity and NAFLD: the role of bacteria and microbiota. Clin Liver Dis 2014;18:59-71.
5 Targher G, Bertolini L, Rodella S, Tessari R, Zenari L, Lippi G, et al. Nonalcoholic fatty liver disease is independently associated with an increased incidence of cardiovascular events in type 2 diabetic patients. Diabetes Care 2007;30:2119-2121.
6 Angulo P. Long-term mortality in nonalcoholic fatty liver disease: is liver histology of any prognostic significance? Hepatology 2010;51:373-375.
7 Bedogni G, Miglioli L, Masutti F, Tiribelli C, Marchesini G, Bellentani S. Prevalence of and risk factors for nonalcoholic fatty liver disease: the Dionysos nutrition and liver study. Hepatology 2005;42:44-52.
8 Ogden CL, Carroll MD, Curtin LR, McDowell MA, Tabak CJ, Flegal KM. Prevalence of overweight and obesity in the United States, 1999-2004. JAMA 2006;295:1549-1555.
9 Clark JM, Brancati FL, Diehl AM. The prevalence and etiology of elevated aminotransferase levels in the United States. Am J Gastroenterol 2003;98:960-967.
10 Vernon G, Baranova A, Younossi ZM. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment Pharmacol Ther 2011;34:274-285.
11 Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology 2004;40:1387-1395.
12 Fischer GE, Bialek SP, Homan CE, Livingston SE, McMahon BJ. Chronic liver disease among Alaska-Native people, 2003-2004. Am J Gastroenterol 2009;104:363-370.
13 Bialek SR, Redd JT, Lynch A, Vogt T, Lewis S, Wilson C, et al. Chronic liver disease among two American Indian patient populations in the southwestern United States, 2000-2003. J Clin Gastroenterol 2008;42:949-954.
14 Day CP, James OF. Steatohepatitis: a tale of two "hits"? Gastroenterology 1998;114:842-845.
15 Dowman JK, Tomlinson JW, Newsome PN. Pathogenesis of non-alcoholic fatty liver disease. QJM 2010;103:71-83.
16 Tilg H, Moschen AR. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology 2010;52:1836-1846.
17 Speliotes EK, Butler JL, Palmer CD, Voight BF; GIANT Consortium; MIGen Consortium; et al. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology 2010;52:904-912.
18 Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology 2009;136:65-80.
19 Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol 2005;3:431-438.
20 Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev 2010;90:859-904.
21 Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science 2006;312:1355-1359.
22 Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science 2005;308:1635-1638.
23 Triantafyllou K, Pimentel M. Small intestinal bacterial overgrowth. In: Lacy BE CM, DiBaise JK (eds), ed. Functional and motility disorders of the gastrointestinal tract. A case study approach. New York: Springer+Bussiness Media; 2015:125-136.
24 Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature 2011;473:174-180.
25 Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 2013;57:601-609.
26 Goel A, Gupta M, Aggarwal R. Gut microbiota and liver disease. J Gastroenterol Hepatol 2014;29:1139-1148.
27 Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480-484.
28 Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol 2013;11:868-875.e1-3.
29 Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 2013;58:120-127.
30 Wong VW, Tse CH, Lam TT, Wong GL, Chim AM, Chu WC, et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis--a longitudinal study. PLoS One 2013;8:e62885.
31 Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 2011;140:976-986.
32 Abu-Shanab A, Quigley EM. The role of the gut microbiota in nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol 2010;7:691-701.
33 Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A 2004;101:15718-15723.
34 Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A 2007;104:979-984.
35 Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature 2007; 449:804-810.
36 Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 2010;18:190-195.
37 Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 2005;102:11070-11075.
38 Kalliomäki M, Collado MC, Salminen S, Isolauri E. Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr 2008;87:534-538.
39 Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature 2006;444:1022-1023.
40 Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A 2009;106:2365-2370.
41 Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105-108.
42 Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucindegrading bacterium. Int J Syst Evol Microbiol 2004;54:1469-1476.
43 Santacruz A, Collado MC, García-Valdés L, Segura MT, Martín-Lagos JA, Anjos T, et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr 2010;104:83-92.
44 Karlsson CL, Onnerfält J, Xu J, Molin G, Ahrné S, Thorngren-Jerneck K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity (Silver Spring) 2012;20:2257-2261.
45 Liou AP, Paziuk M, Luevano JM Jr, Machineni S, Turnbaugh PJ, Kaplan LM. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med 2013;5:178ra41.
46 Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in dietinduced obese mice. Gut 2014;63:727-735.
47 Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 2013;110:9066-9071.
48 Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology 2014;146:1513-1524.
49 Sarkola T, Eriksson CJ. Effect of 4-methylpyrazole on endogenous plasma ethanol and methanol levels in humans. Alcohol Clin Exp Res 2001;25:513-516.
50 Hartmann P, Chen WC, Schnabl B. The intestinal microbiome and the leaky gut as therapeutic targets in alcoholic liver disease. Front Physiol 2012;3:402.
51 Nair S, Cope K, Risby TH, Diehl AM. Obesity and female gender increase breath ethanol concentration: potential implications for the pathogenesis of nonalcoholic steatohepatitis. Am J Gastroenterol 2001;96:1200-1204.
52 Baker SS, Baker RD, Liu W, Nowak NJ, Zhu L. Role of alcohol metabolism in non-alcoholic steatohepatitis. PLoS One 2010;5: e9570.
53 Volynets V, Küper MA, Strahl S, Maier IB, Spruss A, Wagnerberger S, et al. Nutrition, intestinal permeability, and blood ethanol levels are altered in patients with nonalcoholic fatty liver disease (NAFLD). Dig Dis Sci 2012;57:1932-1941.
54 Rao R. Endotoxemia and gut barrier dysfunction in alcoholic liver disease. Hepatology 2009;50:638-644.
55 Sajjad A, Mottershead M, Syn WK, Jones R, Smith S, Nwokolo CU. Ciprofloxacin suppresses bacterial overgrowth, increases fasting insulin but does not correct low acylated ghrelin concentration in non-alcoholic steatohepatitis. Aliment Pharmacol Ther 2005;22:291-299.
56 Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature 2000;406:782-787.
57 Ruiz AG, Casafont F, Crespo J, Cayón A, Mayorga M, Estebanez A, et al. Lipopolysaccharide-binding protein plasma levels and liver TNF-alpha gene expression in obese patients: evidence for the potential role of endotoxin in the pathogenesis of nonalcoholic steatohepatitis. Obes Surg 2007;17:1374-1380.
58 Yang SQ, Lin HZ, Lane MD, Clemens M, Diehl AM. Obesity increases sensitivity to endotoxin liver injury: implications for the pathogenesis of steatohepatitis. Proc Natl Acad Sci U S A 1997;94:2557-2562.
59 Imajo K, Fujita K, Yoneda M, Nozaki Y, Ogawa Y, Shinohara Y, et al. Hyperresponsivity to low-dose endotoxin during progression to nonalcoholic steatohepatitis is regulated by leptinmediated signaling. Cell Metab 2012;16:44-54.
60 Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007;56:1761-1772.
61 Csak T, Velayudham A, Hritz I, Petrasek J, Levin I, Lippai D, et al. Deficiency in myeloid differentiation factor-2 and toll-like receptor 4 expression attenuates nonalcoholic steatohepatitis and fibrosis in mice. Am J Physiol Gastrointest Liver Physiol 2011;300:G433-441.
62 Alisi A, Manco M, Devito R, Piemonte F, Nobili V. Endotoxin and plasminogen activator inhibitor-1 serum levels associated with nonalcoholic steatohepatitis in children. J Pediatr Gastroenterol Nutr 2010;50:645-649.
63 Harte AL, da Silva NF, Creely SJ, McGee KC, Billyard T, Youssef-Elabd EM, et al. Elevated endotoxin levels in nonalcoholic fatty liver disease. J Inflamm (Lond) 2010;7:15.
64 Zhang G, Ghosh S. Molecular mechanisms of NF-kappaB activation induced by bacterial lipopolysaccharide through Tolllike receptors. J Endotoxin Res 2000;6:453-457.
65 Miura K, Kodama Y, Inokuchi S, Schnabl B, Aoyama T, Ohnishi H, et al. Toll-like receptor 9 promotes steatohepatitis by induction of interleukin-1beta in mice. Gastroenterology 2010;139:323-334.e7.
66 Cawthorn WP, Sethi JK. TNF-alpha and adipocyte biology.FEBS Lett 2008;582:117-131.
67 Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 2012;482:179-185.
68 Vance DE. Role of phosphatidylcholine biosynthesis in the regulation of lipoprotein homeostasis. Curr Opin Lipidol 2008;19: 229-234.
69 Zeisel SH, Wishnok JS, Blusztajn JK. Formation of methylamines from ingested choline and lecithin. J Pharmacol Exp Ther 1983;225:320-324.
70 Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U S A 2006;103:12511-12516.
71 Buchman AL, Dubin MD, Moukarzel AA, Jenden DJ, Roch M, Rice KM, et al. Choline deficiency: a cause of hepatic steatosis during parenteral nutrition that can be reversed with intravenous choline supplementation. Hepatology 1995;22:1399-1403.
72 Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C, et al. Intestinal microbiota determines development of nonalcoholic fatty liver disease in mice. Gut 2013;62:1787-1794.
73 Wu WC, Zhao W, Li S. Small intestinal bacteria overgrowth decreases small intestinal motility in the NASH rats. World J Gastroenterol 2008;14:313-317.
74 Fu XS, Jiang F. Cisapride decreasing orocecal transit time in patients with nonalcoholic steatohepatitis. Hepatobiliary Pancreat Dis Int 2006;5:534-537.
75 Miele L, Valenza V, La Torre G, Montalto M, Cammarota G, Ricci R, et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology 2009;49:1877-1887.
76 Sabaté JM, Jouët P, Harnois F, Mechler C, Msika S, Grossin M, et al. High prevalence of small intestinal bacterial overgrowth in patients with morbid obesity: a contributor to severe hepatic steatosis. Obes Surg 2008;18:371-377.
77 Shanab AA, Scully P, Crosbie O, Buckley M, O'Mahony L, Shanahan F, et al. Small intestinal bacterial overgrowth in nonalcoholic steatohepatitis: association with toll-like receptor 4 expression and plasma levels of interleukin 8. Dig Dis Sci 2011;56:1524-1534.
78 Soza A, Riquelme A, González R, Alvarez M, Pérez-Ayuso RM, Glasinovic JC, et al. Increased orocecal transit time in patients with nonalcoholic fatty liver disease. Dig Dis Sci 2005;50:1136-1140.
79 Wigg AJ, Roberts-Thomson IC, Dymock RB, McCarthy PJ, Grose RH, Cummins AG. The role of small intestinal bacterial overgrowth, intestinal permeability, endotoxaemia, and tumour necrosis factor alpha in the pathogenesis of nonalcoholic steatohepatitis. Gut 2001;48:206-211.
80 Gibson GR, Cummings JH, Macfarlane GT, Allison C, Segal I, Vorster HH, et al. Alternative pathways for hydrogen disposal during fermentation in the human colon. Gut 1990;31:679-683.
81 Triantafyllou K, Chang C, Pimentel M. Methanogens, methane and gastrointestinal motility. J Neurogastroenterol Motil 2014;20:31-40.
82 Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A 2006;103:10011-10016.
83 Pimentel M, Mayer AG, Park S, Chow EJ, Hasan A, Kong Y. Methane production during lactulose breath test is associated with gastrointestinal disease presentation. Dig Dis Sci 2003;48:86-92.
84 Basseri RJ, Basseri B, Pimentel M, Chong K, Youdim A, Low K, et al. Intestinal methane production in obese individuals is associated with a higher body mass index. Gastroenterol Hepatol (N Y) 2012;8:22-28.
85 Mathur R, Amichai M, Chua KS, Mirocha J, Barlow GM, Pimentel M. Methane and hydrogen positivity on breath test is associated with greater body mass index and body fat. J Clin Endocrinol Metab 2013;98:E698-702.
86 Mathur R, Kim G, Morales W, Sung J, Rooks E, Pokkunuri V, et al. Intestinal Methanobrevibacter smithii but not total bacteria is related to diet-induced weight gain in rats. Obesity (Silver Spring) 2013;21:748-754.
87 Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 2014;11:506-514.
88 Ritze Y, Bárdos G, Claus A, Ehrmann V, Bergheim I, Schwiertz A, et al. Lactobacillus rhamnosus GG protects against nonalcoholic fatty liver disease in mice. PLoS One 2014;9:e80169.
89 Okubo H, Sakoda H, Kushiyama A, Fujishiro M, Nakatsu Y, Fukushima T, et al. Lactobacillus casei strain Shirota protects against nonalcoholic steatohepatitis development in a rodent model. Am J Physiol Gastrointest Liver Physiol 2013;305:G911-918.
90 Endo H, Niioka M, Kobayashi N, Tanaka M, Watanabe T. Butyrate-producing probiotics reduce nonalcoholic fatty liver disease progression in rats: new insight into the probiotics for the gut-liver axis. PLoS One 2013;8:e63388.
91 Aller R, De Luis DA, Izaola O, Conde R, Gonzalez Sagrado M, Primo D, et al. Effect of a probiotic on liver aminotransferases in nonalcoholic fatty liver disease patients: a double blind randomized clinical trial. Eur Rev Med Pharmacol Sci 2011;15:1090-1095.
92 Vajro P, Mandato C, Licenziati MR, Franzese A, Vitale DF, Lenta S, et al. Effects of Lactobacillus rhamnosus strain GG in pediatric obesity-related liver disease. J Pediatr Gastroenterol Nutr 2011;52:740-743.
93 Malaguarnera M, Vacante M, Antic T, Giordano M, Chisari G, Acquaviva R, et al. Bifidobacterium longum with fructooligosaccharides in patients with non alcoholic steatohepatitis. Dig Dis Sci 2012;57:545-553.
94 Wong VW, Won GL, Chim AM, Chu WC, Yeung DK, Li KC, et al. Treatment of nonalcoholic steatohepatitis with probiotics. A proof-of-concept study. Ann Hepatol 2013;12:256-262.
95 Ma YY, Li L, Yu CH, Shen Z, Chen LH, Li YM. Effects of probiotics on nonalcoholic fatty liver disease: a meta-analysis. World J Gastroenterol 2013;19:6911-6918.
96 Vesper BJ, Jawdi A, Altman KW, Haines GK 3rd, Tao L, Radosevich JA. The effect of proton pump inhibitors on the human microbiota. Curr Drug Metab 2009;10:84-89.
97 Lin EA, Barlow GM, Mathur R. The microbiome in nonalcoholic fatty liver disease: associations and implications. Ann Gastroenterol 2014;27:181-183.
Received February 15, 2015
Accepted after revision June 10, 2015
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Risk factors of metabolic syndrome after liver transplantation
- Combined Hangzhou criteria with neutrophillymphocyte ratio is superior to other criteria in selecting liver transplantation candidates with HBV-related hepatocellular carcinoma
- Warm HTK donor pretreatment reduces liver injury during static cold storage in experimental rat liver transplantation
- Intrahepatic distant recurrence following complete radiofrequency ablation of small hepatocellular carcinoma: risk factors and early MRI evaluation
- Oncogenic role of microRNA-423-5p in hepatocellular carcinoma
- Ankaflavin ameliorates steatotic liver ischemiareperfusion injury in mice
