好氧甲烷氧化菌生态学研究进展
2013-12-09贠娟莉王艳芬张洪勋
贠娟莉,王艳芬,张洪勋
(中国科学院大学,北京 100049)
好氧甲烷氧化菌生态学研究进展
贠娟莉,王艳芬*,张洪勋
(中国科学院大学,北京 100049)
好氧甲烷氧化菌是以甲烷为碳源和能源的细菌。好氧甲烷氧化菌在自然环境中分布广泛,人类已从土壤、淡水和海洋沉积物、泥炭沼泽、热泉、海水和南极环境分离到甲烷氧化菌的纯培养。好氧甲烷氧化菌可分为14个属,包括研究较为深入的隶属于变形菌门Alpha和Gamma纲的细菌,以及属于疣微菌门的极端嗜热嗜酸甲烷氧化菌。最近,好氧甲烷氧化菌还被发现存在于苔藓类植物(尤其是泥炭苔藓)共生体中,兼性营养好氧甲烷氧化菌也被发现。通过对好氧甲烷氧化菌的分类、生理生化特征、分子生物学检测方法以及微生物生态学中的研究成果的总结与分析,以及对甲烷氧化菌研究所面临的问题进行讨论,以期为今后进一步开展好氧甲烷氧化菌及其在碳循环中的作用研究提供参考。
好氧甲烷氧化菌;微生物生态;分类学地位;多样性;碳循环
甲烷是大气中仅次于二氧化碳的第二号温室气体。虽然大气中的甲烷的含量仅为二氧化碳的1/27,但甲烷引起的温室效应是同等质量二氧化碳的20—30倍[1]。造成甲烷浓度升高的主要成因有人为和自然两种因素[2]。人为活动造成的甲烷排放约占总排放量的70%左右(图1),包括水稻种植、垃圾填埋以及生物质燃烧;自然界甲烷排放的主要来源包括自然湿地、植物以及海洋[3]。
大气中甲烷通过数量级相近的源(Source)和汇(Sink)达到平衡,随着大气中甲烷浓度的增加,甲烷的汇也成相应比例增加,从而一定程度上削弱了甲烷源增加造成的环境气候影响,因而实际产生的甲烷量要比测得的排放量大的多[4- 5]。一直以来,甲烷汇的增长小于总甲烷源的增长,从而导致工业革命以来大气中甲烷浓度的持续稳定增长[3]。好氧甲烷氧化菌是重要的甲烷的汇,在环境中起着甲烷生物过滤器的作用,它们能使高达90%由产甲烷古菌所产生的甲烷在排入大气之前被氧化[6- 7]。环境中存在两类截然不同的甲烷氧化菌,好氧甲烷氧化菌(Methanotrophs)和厌氧甲烷氧化菌。
好氧甲烷氧化菌主要存在于甲烷与氧气共存的微小界面空间,包括土壤-空气、水-空气界面、植物根际以及植物内部。大量有关好氧甲烷氧化菌的研究工作显示,该类微生物能适应各种环境。人类对于好氧甲烷氧化菌的研究已较为深入,这些研究不断加深人类对全球甲烷循环的认知。本文就好氧甲烷氧化菌的生理生化及分类学特点、生态学地位以及微生物生态学研究方法进行全面深入的综述,以此阐明好氧甲烷氧化菌在全球碳循环中的重要作用。
1 好氧甲烷氧化菌分类及生理生化特征
1.1 好氧甲烷氧化菌分类
好氧甲烷氧化菌(Aerobic methanotrophs)是以甲烷作为唯一碳源和能源的微生物,是甲基营养细菌(Methylotrophic bacteria)的一个分支[8]。好氧甲烷氧化菌于1906年首次被荷兰微生物学家Sohngen分离出来[9]。1970年Whittenbury等[10]对分离和鉴定的100多种好氧甲烷氧化菌进行了分类,这些分类方法至今仍是鉴定好氧甲烷氧化菌强有力的依据。在Whittenbury等的基础上,Bowman等采用分离方法对更多好氧甲烷氧化菌进行了保存,并进行了更加系统的分类和描述[11- 15]。
已知的好氧甲烷营养菌可分为TypeⅠ型和TypeⅡ型两类,分属于γ-Proteobacteria纲和α-Proteobacteria纲。TypeⅠ型好氧甲烷氧化菌属于Methylococcaceae科,包含Methylobacter,Methylomonas,Methylosoma,Methylomicrobium,Methylococcus,Methylocaldum,Methylothermus,Methylohalobius,Methylosarcina和Methylosphaera10个属。TypeⅠ型好氧甲烷氧化菌中的Methylococcus和Methylocaldum也被称为TypeX型好氧甲烷营养菌,是一类耐热的甲烷氧化菌,其生理生化及系统发育学特征与其他TypeⅠ型甲烷氧化菌有所不同。TypeⅡ型好氧甲烷氧化菌归属于Methylocystaceae和Beijerinckiaceae 2个科,前者包含Methylocystis和Methylosinus属,后者有Methylocapsa和Methylocella属。科学家获得的第1株兼性好氧甲烷氧化菌Methylocellasilvestris为TypeⅡ型甲烷氧化菌。该菌除能利用甲烷外,还能利用多碳化合物作为碳源[16- 17]。后来,两种属于TypeⅠ型好氧甲烷氧化菌的丝状甲烷氧化菌Crenothrixpolyspora[18]和Clonothrixfusca[19]被发现,这2种甲烷氧化菌形成了一个独特的TypeⅠ型甲烷氧化菌分支。值得注意的是,Nature和PNAS杂志报道了3株极端嗜酸嗜热(pH值1.5, 65 ℃)的好氧甲烷氧化菌:Methylokorusinfernorum[20]、Acidimethylosilexfumarolicum[21]和Methyloacidakamchatkensis[22],它们不属于任何一类已知的好氧甲烷氧化菌,而是属于疣微菌门(Verrucomicrobia),研究人员将其命名为Methylacidiphilum属[23]。图1是依据16S rRNA序列绘制的各好氧甲烷氧化菌分支之间的系统进化树。
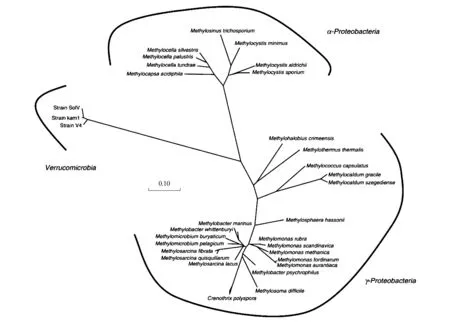
图1 基于16S rRNA序列的好氧甲烷氧化菌进化树[24]Fig.1 16S rRNA gene phylogeny of the aerobic methane oxidizing bacteria
TypeⅠ和TypeⅡ型好氧甲烷氧化菌除系统发育学有各自特点外,在形态学上也具明显差异,TypeⅠ型甲烷氧化菌具有成束的分布于细胞质内的胞质内膜(图2a),而TypeⅡ型甲烷氧化菌一般只含有紧贴外壁的胞质内膜(图2b)[24]。
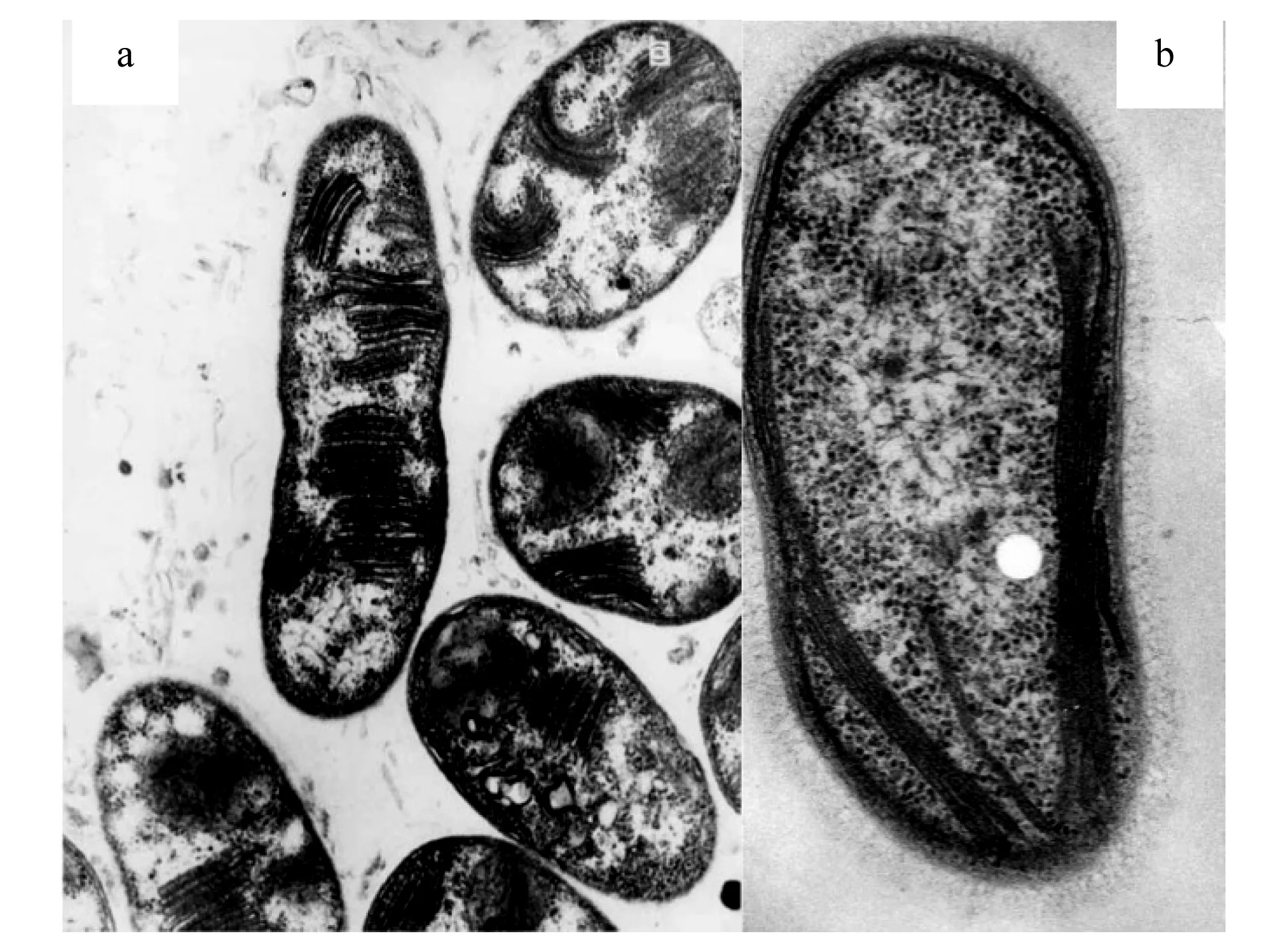
图2 TypeⅠ型好氧甲烷氧化菌代表菌Methylomonas methanica电镜照片(a);TypeⅡ型好氧甲烷氧化菌代表菌Methylosinus trichosporium电镜照片(b)[24]Fig.2 Electron micrograph of a cross-section of a typical TypeⅠ methanotroph Methylomonas methanica (a); Electron micrograph of a cross-section of a typical TypeⅡ methanotroph Methylosinus trichosporium (b)[24]
1.2 好氧甲烷氧化菌生理生化特征
环境中由好氧甲烷氧化菌推动的甲烷氧化主要过程为:好氧甲烷氧化菌首先利用自身携带的甲烷单加氧酶(Methane monooxygenase, MMO)催化甲烷氧化为甲醇,甲醇接着被甲醇脱氢酶催化氧化生成甲醛。最后,好氧甲烷氧化菌通过丝氨酸途径(Serine pathway)或单磷酸核酮糖途径(RuMP pathway)将甲醛转化为细胞物质[25]。
好氧甲烷氧化菌中存在2种甲烷单加氧酶:一种是与细胞膜结合,含铜、铁离子的颗粒性甲烷单加氧酶(Particulate methane monooxygenase,pMMO),存在于除Methylocella[26]及Methyloferula[27]以外的所有已发现的好氧甲烷氧化菌中;另一种是分泌在周质空间中的可溶性甲烷单加氧酶(Soluble methane monooxygenase,sMMO),存在于部分甲烷氧化菌中。由于MMO是甲烷氧化菌的功能酶系,且几乎所有好氧甲烷氧化菌都含有pMMO,因此利用MMO(尤其是pmoA,编码pMMO的一段基因)作为生物标记物进行好氧甲烷氧化菌生态学研究已广为采用。部分好氧甲烷氧化菌,如所有TypeⅡ型甲烷氧化菌、TypeⅠ型甲烷氧化菌的Methylomonas、Methylobacter和Methylococcus属,除有氧化甲烷能力外,还有固氮能力。因此,利用nifH基因也可对该类好氧甲烷氧化菌进行分子生态学研究[28]。对于MMO的应用研究主要集中在两方面:一是通过基因突变等手段,对好氧甲烷氧化菌编码MMO等蛋白的基因进行改造,从而满足不同工业催化的需要,例如提高MMO的活性、改变MMO的底物范围、提高其对金属离子的耐受性等;二是通过代谢工程手段,向好氧甲烷氧化菌内部引入外源基因,利用甲烷氧化菌为载体生产高附加值的工业产品,如表达生产蛋白等生物产品[29]。
2 好氧甲烷氧化菌分子生态学研究方法
传统方法是利用NMS及ANMS等无机盐培养基对好氧甲烷氧化菌进行富集培养或者菌株分离[10]。分子微生物生态学方法的出现及应用极大扩展了人类对甲烷氧化菌的认知范围。
最常用的好氧甲烷氧化菌的分子标记物是16S rRNA基因,该项应用主要基于大量的16S rRNA基因数据库。针对好氧甲烷氧化菌各属或菌株的特异性引物和探针已有大量报道,这些引物与以PCR技术为基础的克隆文库(Clone library)、变形梯度凝胶电泳(DGGE)、荧光原位杂交(FISH)等分析技术相结合,在环境微生物生态学研究中发挥重要作用[30]。但也会由于引物特异性不足,从而造成非特异性扩增,因此在以16S rRNA基因为对象研究环境中好氧甲烷氧化菌时需考虑到这一因素。除了16S rRNA基因之外,功能基因也是研究环境中好氧甲烷氧化菌的强有力工具,这些功能功能基因包括mmoX、pmoA、mxaF及nifH[28]。
DGGE和末端限制多态性研究(T-RFLP)为对比大量环境样品中甲烷氧化菌多样性差异提供了快速、灵敏的技术。许多针对这2种方法设计的16S rRNA基因和功能基因引物为研究环境中好氧甲烷氧化菌多样性提供了强有力的工具[30]。另外一种研究环境中甲烷氧化菌的高通量方法是生物芯片技术,尽管生物芯片最初是作为基因组表达分析的研究工具,但基因诊断芯片已成功开发并已应用于环境中好氧甲烷氧化菌的检测[31]。为了定量环境中好氧甲烷氧化菌的数量,可培养方法(最大释然法,即MPN法)和不依赖培养的分子生物学方法(Real-time PCR和FHSH)均被使用[30]。这2种方法各有利弊,MPN技术依赖于特定培养基中甲烷氧化菌的生长情况,具有很大的偏好性;分子生物学技术虽不需培养,但很大程度上取决于环境样品的类型和质量好坏。为了检测研究环境样品中活跃的甲烷氧化菌菌群,稳定同位素探针技术(SIP)应运而生。这项应用技术包括DNA-SIP[32]、RNA-SIP[33]、磷脂脂肪酸(PLFA)-SIP[34]以及最新使用的mRNA-SIP[35]。除此外,SIP技术也和宏基因组学相结合用于发现新的好氧甲烷氧化菌[36]。除了以上常用的分子生态学研究方法外,其他研究工具也逐渐被引入环境好氧甲烷氧化菌的研究,例如显微镜放射自显影(MAR)-FISH[37- 38]、同位素芯片[39]、Raman-FISH[40]、纳米二次离子质谱(NanoSIMS)[41]和微流体数字PCR[42]。这些技术检出限更高、可同时检测多个样品且能直接给出所测定菌株或菌群的功能特征。
3 好氧甲烷氧化菌的生态学研究
过去几十年中,培养及不依赖培养的分子生态学方法已经被广泛用于各种环境中好氧甲烷氧化菌的多样性、分布及丰度研究,如稻田、垃圾填埋厂、淡水和淡水沉积物、海水、山地土壤以及极端环境。
稻田是大气甲烷的主要来源之一,全球人口激增导致大米需求增加,故而稻田甲烷排放量呈增加趋势。研究表明稻田中好氧甲烷氧化菌多样性较高[43],包括Methylomonas、Methylobacter、Methylomicrobium、Methylococcus、Methylocaldum、Methylocystis和Methylosinus属。有关稻田中何种好氧甲烷氧化菌占据优势,各地研究结果并不一致。陆雅海等经研究发现,水稻根部对TypeⅠ型好氧甲烷氧化菌具有选择性,且水稻根部比根际土壤中TypeⅠ型好氧甲烷氧化菌更为丰富[44]。另有报道认为水稻根系对TypeⅠ型好氧甲烷氧化菌的偏好不受水稻物种的影响。除Methylocaldum属的好氧甲烷氧化菌多在热带地区被发现外,稻田中的好氧甲烷氧化菌在全球范围内并没有明显的地域性特征。研究者认为TypeⅠ型好氧甲烷氧化菌成为水稻根际优势菌群的原因在于其能适应较广的O2/CH4范围。再者,水稻根系的O2浓度非常不稳定,不适宜TypeⅡ型好氧甲烷氧化菌的生长[45]。与以上结果相反,Luke等人通过T-RFLP和基因诊断芯片技术对18种不同水稻品种进行研究,发现这些水稻根系中以TypeⅡ和TypeX型好氧甲烷氧化菌为主,并显示出极大的多样性,该研究小组还指出水稻根部的好氧甲烷氧化菌群落组成受水稻基因型影响很大[46]。郑勇等[47]研究发现TypeⅡ型甲烷氧化菌在长期施肥的水稻土壤中占优势,定量PCR结果发现所有处理中TypeⅡ型甲烷氧化菌的数量是TypeⅠ型好氧甲烷氧化菌的1.88至3.32倍。不同施肥处理对甲烷氧化菌的菌群组成有所影响。长期施氮磷钾和秸秆的处理(NPK+C)、施氮肥和钾肥(NK)处理的稻田土壤中TypeⅡ型甲烷氧化菌的数量明显比对照高,表明长期施肥对TypeⅡ型甲烷氧化菌的生长有促进作用。稻田中甲烷氧化菌的分布和丰度受很多因素的影响,如氧气的可用性及水稻的生长时期等因素[48]。在稻田土壤中,高氧气、低甲烷的环境利于TypeⅠ型好氧甲烷氧化菌的生长,反之则利于TypeⅡ型甲烷氧化菌的生长[49]。Shrestha等研究发现,无论施肥与否,在水稻各个生长阶段,根际土壤中好氧甲烷氧化菌以TypeⅡ型为主,而水稻根部则以TypeⅠ型为主[50]。

淡水和淡水沉积物是大气甲烷的又一重要来源,该类环境中好氧甲烷氧化菌主要以TypeⅠ型甲烷氧化菌中的Methylomonas、Methylobacter、Methylosarcina、Methylococcus和Methylosoma属为主。在对华盛顿湖沉积物的研究中发现,TypeⅠ型比TypeⅡ型好氧甲烷氧化菌多1—2个数量级[59]。另外,在康士坦茨湖中,TypeⅠ型甲烷氧化菌占pmoA的克隆文库序列的90%[60]。Dumont等利用DNA-SIP和mRNA-SIP相结合的方法,发现在Stechlin湖中也以TypeⅠ型甲烷氧化菌为主要菌群[35]。张洪勋等通过对我国两处淡水沼泽湿地研究发现:我国青藏高原若尔盖永冻土湿地中好氧甲烷氧化菌仅有Methylobacter属和Methylocystis两个属,且以TypeⅠ型甲烷氧化菌为优势菌群,不同植被覆盖的泥炭沼泽中好氧甲烷氧化菌数量有所不同[61- 62];对我国东北地区松嫩平原向海湿地中好氧甲烷氧化菌多样性进行研究,发现向海湿地好氧甲烷氧化菌多样性与淡水湖泊相似,较若尔盖高寒湿地种类多,但仍以TypeⅠ型甲烷氧化菌的Methylobacter属为优势菌群[63]。研究的两个湿地中均有与Methylococcus属甲烷氧化菌相似度较高的新的甲烷氧化菌存在。另外,这2个湿地中Methylobacter属的甲烷氧化菌亲缘关系相近,表明我国自然湿地中甲烷氧化菌具有地域性特点。
对海水和海洋沉积物中甲烷氧化菌的研究相对较少,虽然从海水中分离到了Methylomonas和Methylomicrobium属的甲烷氧化菌,但分子生态学方面的研究却证明不可培养的好氧甲烷氧化菌Methylococcaceae科的甲烷氧化菌在海洋水体中占主导地位[64]。还有研究发现OPU1, OPU3和Group X是深海中占主导地位的好氧甲烷氧化菌,其中OPU1和OPU3菌群在墨西哥湾和Santa Monica海湾具有生长优势,且其在甲烷溪流的数量比非甲烷溪流中多50多倍,Group X则在加利福尼亚海岸中占优势,其生长不受甲烷溪流的影响[65]。在黑海浅海中发现了好氧甲烷氧化菌,TypeⅠ和Ⅱ型好氧甲烷氧化菌各占细菌总数的2.5%,在深海中参与甲烷氧化的菌群则主要是ANME-1和ANME-2厌氧甲烷氧化菌[66]。
山地和森林是大气甲烷主要的汇,这些环境中的好氧甲烷氧化菌对大气甲烷有很高的亲和力,并以TypeⅡ型的Methylocystis属,山地土壤菌群α(USCα)和γ(USCγ)的甲烷氧化菌为主。Kolb等[67]发现“USCα”型甲烷氧化菌是酸性森林土壤中的主要菌群,而“Cluster I”则在中性森林土壤中占优势,研究者认为这两种甲烷氧化菌能适应低甲烷浓度环境主要是由于其细胞特异性的甲烷氧化能力。Horz等[68]在加利福尼亚山地土壤中发现3个不同的甲烷氧化菌分支,这些分支与已报道的RA14或VB5FH-A种群类似,这些甲烷氧化菌菌群是典型的大气甲烷氧化菌群,其对气候变暖的响应程度与TypeⅡ型甲烷氧化菌不同。Mohanty等[69]研究发现,森林中的甲烷氧化菌以TypeⅡ型的Methylocystis属为主,且有TypeⅠ型的Methylomicrobium和Methylosarcina属甲烷氧化菌存在。Menyailo等[70]对西伯利亚森林中不同树种的土壤中好氧甲烷氧化菌的菌群组成及丰度进行了调查,发现该地土壤中以USCα为主要甲烷氧化菌群,树种的不同并不会影响甲烷氧化菌的菌群组成,但会影响土壤甲烷氧化速率,造成这一结果的原因可能是树种的不同会影响甲烷氧化菌的单个细胞活性,但并不会影响其周围土壤中甲烷氧化菌的种类,这一过程是通过控制土壤中氮循环完成的。有研究者推测,USCα和USCγ为不可培养的可在大气低浓度甲烷下生长的甲烷氧化菌。另一种解释则认为在高山和森林中的好氧甲烷氧化菌以木质素降解产物甲醇作为能源和碳源。直到近期,Baani和Liesack[71]发现在Methylocystissp. SC2中存在一种特殊的pMMO酶,该酶对甲烷的亲和力不同,由此可解释为什么Methylocystis属的甲烷氧化菌能在山地和森林,以及其他环境中普遍存在。
极端环境中的好氧甲烷氧化菌研究一直备受关注。从酸性从泥炭沼泽土壤和酸性森林土壤中分离出的属于Methylocella和Methylocapsa属的好氧甲烷氧化菌[72- 73];从嗜盐和嗜碱环境中分离到的属于Methylomicrobium属和Methylohalobius的好氧甲烷氧化菌[74- 75];从永冻土地区分离出的嗜冷甲烷氧化菌Methylobacterpsychrophilus、Methylosphaerahansonii和Methylomonasscandinavica[13, 76],这些TypeⅠ型甲烷氧化菌生长于低温环境中(5—15 ℃)并且G+C含量较低;以及从热泉中分离的嗜热甲烷氧化菌MethylococcuscapsulatusBath[10]、Methylocaldumspp.[77- 78]和Methylothermusthermalis[79]。最为引人瞩目的是从世界不同区域火分离的疣微菌门(Verrucomicrobia)的3株极端嗜热嗜酸甲烷氧化菌[20- 22],这些好氧甲烷氧化菌细胞内含有与其他菌株不同的pmoA基因,表明它们对碳的代谢和吸收可能有另外的途径。对于疣微菌门好氧甲烷氧化菌物理化学特征及分类学地位,Op den Camp等人给予了详细的综述[23]。人类对极端环境中好氧甲烷氧化菌的探索有待进一步深入,相信将来还会有更多极端环境中的好氧甲烷氧化菌被发现。
4 苔藓-好氧甲烷氧化菌共生体
很长一段时间内,人们认为只有厌氧甲烷氧化菌能够与其他生物形成共生体系,如厌氧甲烷氧化菌和硫还原细菌或硝酸还原菌共生。最新研究发现,好氧甲烷氧化菌也能与环境中的其他生物形成共生体系。Kip与同事研究发现,泥炭苔藓可与好氧甲烷氧化菌共生,合作进行甲烷氧化(图3)[80]。
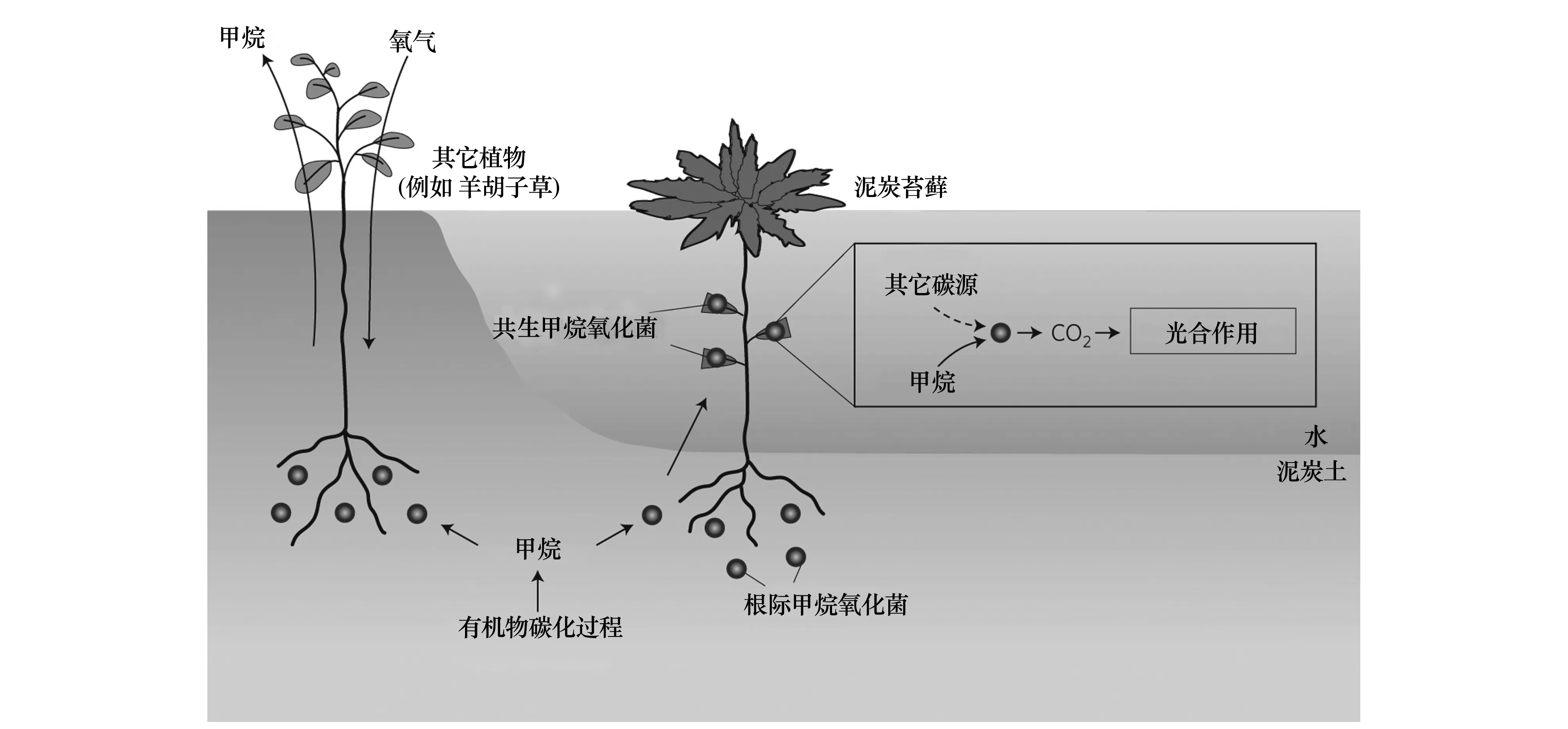
图3 泥炭沼泽中泥炭苔藓-甲烷氧化菌共生甲烷氧化途径示意图[80]Fig.3 Methane oxidation by methanotrophs in a peat bog. Sphagnum mosses form symbioses with methane-consuming bacteria in Sphagnum-dominated peat bogs[80]
稳定同位素标记实验证实了泥炭苔藓所同化的35%的CO2是通过甲烷氧化产生。这种共生体系的形成对于好氧甲烷氧化菌和苔藓来说是互利的(图3):水下的泥炭苔藓由于缺乏气孔不能从大气得到足够的CO2进行光合作用,于是好氧甲烷氧化菌的代谢产物CO2就成为其绝佳的CO2来源。与此同时,水下泥炭苔藓中的好氧甲烷氧化菌也可利用泥炭苔藓光合作用产生的O2完成甲烷氧化过程。Kip等人还指出,在温度升高时,该共生体系能更好的减少甲烷排放。当从土壤中去掉泥炭苔藓时,土壤的甲烷排放量升至原来的4倍,表明泥炭苔藓-好氧甲烷氧化菌共生体系比游离生长的甲烷氧化菌在甲烷氧化过程中起更重要的作用[80- 81]。
通过进一步研究,Kip等人从泥炭苔藓中分离出了多株嗜酸性甲烷氧化菌,所获多数菌株属于TypeⅡ型好氧甲烷氧化菌[82]。另外,他们还在荷兰及巴塔哥尼亚泥炭苔藓中发现Methylomonas和Methylocystis分布最为广泛[82- 83]。Liebner等人[84]通过研究证实,苔藓-甲烷氧化菌共生氧化甲烷的现象不仅存在于泥炭苔藓和低pH值的泥炭沼泽中,还存在于褐藓与永冻土中。该研究同样通过稳定同位素方法研究了西伯利亚冻土水体中的褐藓-好氧甲烷氧化菌共生体系,发现该体系氧化甲烷能力在淹水条件下增强,且在光照条件下是一个纯的甲烷汇,当去掉光照时,土壤就变成了强大的甲烷排放源。该研究还估测,褐藓-好氧甲烷氧化菌共生作用使得北极多边苔原冻土甲烷排放总量至少减少了5%,鉴于褐藻在永冻土淡水区域广泛生长,苔藓-甲烷氧化菌共生作用可能是该地区淡水环境主要的甲烷氧化途径。
5 环境对好氧甲烷氧化菌的影响
影响甲烷氧化菌的环境因素可归为化学因素和生物因素两大类。化学因素主要包括氧气,水分状况,含氮化合物及重金属等;生物因素主要有竞争和捕食。目前研究主要集中在化学因素方面。水位和氧气会影响好养甲烷氧化菌的活性,尤其是在泥炭沼泽湿地和稻田中。有研究证明长期排水会影响土壤中甲烷氧化菌的群落组成,不同形式的土地利用显著影响甲烷氧化菌的群落组成与活性[85]。一方面,水位升高会降低土壤氧气浓度,从而导致甲烷氧化菌可用氧气量减少;另一方面,排水会增加土壤氧气含量从而促进甲烷氧化菌生长。

生物因素也会影响环境中好氧甲烷氧化菌对甲烷的氧化,如捕食。Murase和Frenze[89]做了一项有趣的研究,发现原生动物对甲烷氧化菌的捕食行为,他们从稻田中分离出了以好氧甲烷氧化菌为食的原生动物。Murase等人还发现原生动物偏好捕食特定的甲烷氧化菌(如Methylobacter),这可能是由于捕食这类甲烷氧化菌后能最快速有效的进行自身的同化作用[90]。Moon等将含有蚯蚓的水稻土作为垃圾填埋场的覆盖层能有效减少垃圾填埋场的甲烷排放,并且发现覆盖层土壤中的细菌及甲烷氧化菌主要来源于水稻土及蚯蚓排泄物,TypeⅠ (主要为Methylocaldum)和TypeⅡ (主要为Methylocystis)型甲烷氧化菌都在甲烷氧化过程中起重要作用[91]。近期研究还表明在土壤中加入蚯蚓后能显著增加好氧甲烷氧化菌的多样性和数量[92]。
放牧对甲烷氧化菌有一定影响。周小奇等对我国青藏高原草甸土壤研究发现,放牧显著影响土壤中好氧甲烷氧化菌的群落组成[93]。郑勇等研究发现,放牧增加了好氧甲烷氧化菌的数量,从而促进了土壤的甲烷氧化能力[94]。
6 问题与展望
人类对好氧甲烷氧化菌及其在减少大气甲烷排放的作用研究历经数十载,逐渐阐明了其在大气碳循环中的重要作用,但研究过程中仍涉及到几个重要问题仍值得深思,有待进一步研究[24]。
(1) 好氧甲烷氧化菌多样性及分布在微生物世界内究竟是怎样?过去几年中不断有新的甲烷氧化菌菌株分离出来,如丝状甲烷氧化菌Crenothrix以及嗜酸嗜低温的疣微菌门(Verrucomicrobia)好氧甲烷氧化菌的发现。这些新的发现令人不禁想到,环境中或许还存在更多未知的、新的好氧甲烷氧化菌有待发现。另外,是否存在一类“大气好氧甲烷氧化菌”?是否有好氧甲烷氧化古菌的存在?所有这些问题的解决都将取决于新的分离技术的出现。
(2) 不同种类的好氧甲烷氧化菌菌群之间是怎样相互竞争最基本的生存物质,如氧气和氮素?随着新近发明的可测定单个细胞的Raman荧光原位杂交及纳米二次离子质谱(NanoSIMS)技术的问世,相信这一问题也会迎刃而解。
(3) 兼性甲烷氧化菌是否只存在于Methylocella属,如果不是,那么还有哪些?它们何时以及怎样从异养型微生物转换为甲烷营养型的?作为兼性营养的好氧甲烷氧化有哪些生长优势?比较基因组学和蛋白质组学在定义代谢途径及兼性营养的基因调控机制方面具有重要作用,这些研究的应用将为以上问题的解决带来可能。
[1] IPCC. Climate Change 2007: The physical science basis. Summary for policymakers. Contribution of Working Group I to the fourth assessment report of the Intergovernmental Panel on Climate Change. Paris: Summary for Policymakers formally approved at the 10th Session of Working Group I of the IPCC.
[2] Blake D R, Rowland F S. Continuing worldwide increase in tropospheric methane, 1978 to 1987. Science, 1988, 239(4844): 1129- 1131.
[3] Conrad R. The global methane cycle: recent advances in understanding the microbial processes involved. Environmental Microbiology Reports, 2009, 1(5): 285- 292.
[4] Frenzel P. Plant-associated methane oxidation in rice fields and wetlands // Bernhard S, ed. Advances in Microbial Ecology. New York: Kluwer Academic/Plenum Publisher, 2000: 85- 114.
[5] Reeburgh W S. Global methane biogeochemistry. Treatise on Geochemistry, 2003, 4: 65- 89.
[6] Roslev P, King G M. Regulation of methane oxidation in a freshwater wetland by water table changes and anoxia. FEMS Microbiology Ecology, 1996, 19(2): 105- 115.
[7] Le Mer J, Roger P. Production, oxidation, emission and consumption of methane by soils: a review. European Journal of Soil Biology, 2001, 37(1): 25- 50.
[8] Hanson R S, Hanson T E. Methanotrophic bacteria. Microbiology and Molecular Biology Reviews, 1996, 60(2): 439- 71.
[9] Söhngen N L. Über Bakterien welche methan kohlenstoffnahrung energiequelle gebrauchen. Zentrabl Bakteriol Parasitenkd Infectionskr, 1906, 15: 513- 517.
[10] Whittenbury R, Phillips K C, Wilkinson J F. Enrichment, isolation and some properties of methane-utilizing bacteria. Journal of General Microbiology, 1970, 61(2): 205- 218.
[11] Bowman J P. The methanotrophs-the families Methylococcaceae and Methylocystaceae. Prokaryotes, 2006, 5: 266- 289.
[12] Bowman J P. The methanotrophs- the families Methylococcaceae and Methylocystaceae // Dworkin D M, ed. The Prokaryotes. New York: Springer, 1999: 1953- 1966.
[13] Bowman J P, McCammon S A, Skerratt J H.Methylosphaerahansoniigen. nov., sp. nov., a psychrophilic, group I methanotroph from Antarctic marine-salinity, meromictic lakes. Microbiology, 1997, 143(4): 1451- 1459.
[14] Bowman J P, Sly L I, Nichols P D, Hayward A C. Revised taxonomy of the Methanotrophs: Description ofMethylobactergen. nov., Emendation ofMethylococcus, Validation ofMethylosinusandMethylocystisSpecies, and a proposal that the family Methylococcaceae includes only the group-I Methanotrophs. International Journal of Systematic Bacteriology, 1993, 43(4): 735- 753.
[15] Bowman J P, Sly L I, Stackebrandt E. The phylogenetic position of the family Methylococcaceae. International Journal of Systematic Bacteriology, 1995, 45(1): 182- 185.
[16] Dedysh S N, Knief C, Dunfield P F. Methylocella species are facultatively methanotrophic. Journal of Bacteriology, 2005, 187(13): 4665- 4670.
[17] Theisen A R, Ali M H, Radajewski S, Dumont M G, Dunfield P F, McDonald I R, Dedysh S N, Miguez C B, Murrell J C. Regulation of methane oxidation in the facultative methanotrophMethylocellasilvestrisBL2. Molecular Microbiology, 2005, 58(3): 682- 692.
[18] Stoecker K, Bendinger B, Sch ning B, Nielsen P H, Nielsen J L, Baranyi C, Toenshoff E R, Daims H, Wagner M. Cohn′sCrenothrixis a filamentous methane oxidizer with an unusual methane monooxygenase. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(7): 2363- 2367.
[19] Kulichevskaya I S, Ivanova A O, Belova S E, Baulina O I, Bodelier P L E, Rijpstra W I C, Sinninghe Damsté J S, Zavarzin G A, Dedysh S N. Schlesneriapaludicolagen. nov., sp. nov., the first acidophilic member of the orderPlanctomycetales, fromSphagnum-dominated boreal wetlands. International Journal of Systematic and Evolutionary Microbiology, 2007, 57(11): 2680- 2687.
[20] Dunfield P F, Yuryev A, Senin P, Smirnova A V, Stott M B, Hou S B, Ly B, Saw J H, Zhou Z M, Ren Y, Wang J M, Mountain B W, Crowe M A, Weatherby T M, Bodelier P L E, Liesack W, Feng L, Wang L, Alam M. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature, 2007, 450(7171): 879- 82.
[21] Pol A, Heijmans K, Harhangi H R, Tedesco D, Jetten M S, Op den Camp H J M. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature, 2007, 450(7171): 874- 878.
[22] Islam T, Jensen S, Reigstad L J, Larsen Ø, Birkeland N K. Methane oxidation at 55℃ and pH 2 by a thermoacidophilic bacterium belonging to theVerrucomicrobiaphylum. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(1): 300- 304.
[23] Op den Camp H J M, Islam T, Stott M B, Harhangi H R, Hynes A, Schouten S, Jetten M S M, Birkeland N K, Pol A, Dunfield P F. Environmental, genomic and taxonomic perspectives on methanotrophicVerrucomicrobia. Environmental Microbiology Reports, 2009, 1(5): 293- 306.
[24] Murrell J C. The aerobic methane oxidizing bacteria (Methanotrophs) // Timmis K N, ed. Handbook of Hydrocarbon and Lipid Microbiology. Berlin Heidelberg: Springer, 2010: 1954- 1963.
[25] Mancinelli R L. The regulation of methane oxidation in soil. Annual Review of Microbiology, 1995, 49(1): 581- 605.
[26] Dedysh S N, Liesack W, Khmelenina V N, Suzina N E, Trotsenko Y A, Semrau J D, Bares A M, Panikov N S, Tiedje J M. Methylocella palustris gen. nov., sp nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. International Journal of Systematic and Evolutionary Microbiology, 2000, 50(3): 955- 969.
[27] Vorobev A V, Baani M, Doronina N V, Brady A L, Liesack W, Dunfield P F, Dedysh S N.Methyloferulastellatagen. nov., sp nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. International Journal of Systematic and Evolutionary Microbiology, 2011, 61(10): 2456- 2463.
[28] Trotsenko Y A, Murrell J C. Metabolic aspects of aerobic obligate methanotrophy// Laskin A I, Gadd G M, Sariaslani S, eds. Advances in Applied Microbiology. New York: Academic Press, 2008: 183- 229.
[29] Han B, Su T, Li X, Xing X H. Research progresses of methanotrophs and methane monooxygenases. Chinese Journal of Biotechnology, 2008, 24(9): 1511- 1519.
[30] McDonald I R, Bodrossy L, Chen Y, Murrell J C. Molecular ecology techniques for the study of aerobic methanotrophs. Applied and Environmental Microbiology, 2008, 74(5): 1305.
[31] Bodrossy L, Stralis-Pavese N, Murrell J C, Radajewski S, Weilharter A, Sessitsch A. Development and validation of a diagnostic microbial microarray for methanotrophs. Environmental Microbiology, 2003, 5(7): 566- 582.
[32] Radajewski S, Ineson P, Parekh N R, Murrell J C. Stable-isotope probing as a tool in microbial ecology. Nature, 2000, 403(6770): 646- 649.
[33] Manefield M, Whiteley A S, Griffiths R I, Bailey M J. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Applied and Environmental Microbiology, 2002, 68(11): 5367- 5373.
[34] Bull I D, Parekh N R, Hall G H, Ineson P, Evershed R P. Detection and classification of atmospheric methane oxidizing bacteria in soil. Nature, 2000, 405(6783): 175- 178.
[35] Dumont M G, Pommerenke B, Casper P, Conrad R. DNA-, rRNA- and mRNA-based stable isotope probing of aerobic methanotrophs in lake sediment. Environmental Microbiology, 2011, 13(5): 1153- 1167.
[36] Chen Y, Dumont M G, Neufeld J D, Bodrossy L, Stralis‐Pavese N, McNamara N P, Ostle N, Briones M J I, Murrell J C. Revealing the uncultivated majority: combining DNA stable‐isotope probing, multiple displacement amplification and metagenomic analyses of uncultivatedMethylocystisin acidic peatlands. Environmental Microbiology, 2008, 10(10): 2609- 2622.
[37] Lee N, Nielsen P H, Andreasen K H, Juretschko S, Nielsen J L, Schleifer K H, Wagner M. Combination of fluorescent in situ hybridization and microautoradiography-a new tool for structure-function analyses in microbial ecology. Applied and Environmental Microbiology, 1999, 65(3): 1289- 1297.
[38] Ouverney C C, Fuhrman J A. Combined microautoradiography- 16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Applied and Environmental Microbiology, 1999, 65(4): 1746- 1752.
[39] Adamczyk J, Hesselsoe M, Iversen N, Horn M, Lehner A, Nielsen P H, Schloter M, Roslev P, Wagner M. The isotope array, a new tool that employs substrate-mediated labeling of rRNA for determination of microbial community structure and function. Applied and Environmental Microbiology, 2003, 69(11): 6875- 6887.
[40] Huang W E, Stoecker K, Griffiths R, Newbold L, Daims H, Whiteley A S, Wagner M. Raman‐FISH: combining stable‐isotope Raman spectroscopy and fluorescenceinsituhybridization for the single cell analysis of identity and function. Environmental Microbiology, 2007, 9(8): 1878- 1889.
[41] Li T L, Wu T D, Mazéas L, Toffin L, Guerquin‐Kern J L, Leblon G, Bouchez T. Simultaneous analysis of microbial identity and function using NanoSIMS. Environmental Microbiology, 2008, 10(3): 580- 588.
[42] Hashsham S A, Gulari E, Tiedje J M. Microfluidic systems being adapted for microbial, molecular biological analyses. Microbe-American Society for Microbiology, 2007, 2(11): 531- 536.
[43] Hoffmann T, Horz H P, Kemnitz D, Conrad R. Diversity of the particulate methane monooxygenase gene in methanotrophic samples from different rice field soils in China and the Philippines. Systematic and Applied Microbiology, 2002, 25(2): 267- 274.
[44] Wu L Q, Ma K, Lu Y H. Rice roots select for type I methanotrophs in rice field soil. Systematic and Applied Microbiology, 2009, 32(6): 421- 428.
[45] Lüke C, Krause S, Cavigiolo S, Greppi D, Lupotto E, Frenzel P. Biogeography of wetland rice methanotrophs. Environmental Microbiology, 2010, 12(4): 862- 872.
[46] Lüke C, Bodrossy L, Lupotto E, Frenzel P. Methanotrophic bacteria associated to rice roots: the cultivar effect assessed by T-RFLP and microarray analysis. Environmental Microbiology Reports, 2011, 3(5): 518- 525.
[47] Zheng Y, Zhang L M, Zheng Y M, Di H J, He J Z. Abundance and community composition of methanotrophs in a Chinese paddy soil under long-term fertilization practices. Journal of Soils and Sediments, 2008, 8(6): 406- 414.
[48] Eller G, Stubner S, Frenzel P. Group-specific 16S rRNA targeted probes for the detection of type I and type II methanotrophs by fluorescence in situ hybridisation. FEMS Microbiology Letters, 2001, 198(2): 91- 97.
[49] Graham D W, Chaudhary J A, Hanson R S, Arnold R G. Factors affecting competition between TypeⅠ and TypeⅡ methanotrophs in two-organism, continuous-flow reactors. Microbial Ecology, 1993, 25(1): 1- 17.
[50] Shrestha M, Shrestha P M, Frenzel P, Conrad R. Effect of nitrogen fertilization on methane oxidation, abundance, community structure, and gene expression of methanotrophs in the rice rhizosphere. ISME Journal, 2010, 4(12): 1545- 1556.
[51] Howeling S, Kaminski T, Dentener F, Lelieveld J, Heimann M. Inverse modeling of methane sources and sinks using the adjoint of a global transport model. Jounal of Geophysical Research, 1999, 104(D21): 26137- 26160.
[52] Whalen S C, Reeburgh W S, Sandbeck K A. Rapid methane oxidation in a landfill cover soil. Applied and Environmental Microbiology, 1990, 56(11): 3405- 3411.
[53] Jones H A Nedwell D B. Methane emission and methane oxidation in landfill cover soil. FEMS Microbiology Ecology, 1993, 102(3/4): 185- 195.
[54] Hilger H Humer M. Biotic landfill cover treatments for mitigating methane emissions. Environmental Monitoring and Assessment, 2003, 84(1/2): 71- 84.
[55] Chen Y, Murrell J C. Ecology of aerobic methanotrophs and their role in methane cycling // Timmis K N, ed. Handbook of Hydrocarbon and Lipid Microbiology. Berlin Heidelberg: Springer, 2010: 3067- 3076.
[56] Cébron A, Bodrossy L, Chen Y, Singer A C, Thompson I P, Prosser J I, Murrell J C. Identity of active methanotrophs in landfill cover soil as revealed by DNA-stable isotope probing. FEMS Microbiology Ecology, 2007, 62(1): 12- 23.
[57] Chen Y, Dumont M G, Cébron A, Murrell J C. Identification of active methanotrophs in a landfill cover soil through detection of expression of 16S rRNA and functional genes. Environmental Microbiology, 2007, 9(11): 2855- 2869.
[58] Yang N, Lü F, He P J, Shao L M. Response of methanotrophs and methane oxidation on ammonium application in landfill soils. Applied Microbiology and Biotechnology, 2011, 92(5): 1073- 1082.
[59] Costello A M, Auman A J, Macalady J L, Scow K M, Lidstrom M E. Estimation of methanotroph abundance in a freshwater lake sediment. Environmental Microbiology, 2002, 4(8): 443- 450.
[60] Rahalkar M Schink B. Comparison of aerobic methanotrophic communities in littoral and profundal sediments of Lake Constance by a molecular approach. Applied and Environmental Microbiology, 2007, 73(13): 4389- 4394.
[61] Yun J L, Zhuang G Q, Ma A Z, Guo H G, Wang Y F, Zhang H X. Community structure, abundance, and activity of methanotrophs in the Zoige Wetland of the Tibetan Plateau. Microbial Ecology, 2012, 63(4): 835- 843.
[62] Yun J L, Ma A Z, Li Y M, Zhuang G Q, Wang Y F, Zhang H X. Diversity of methanotrophs in Zoige wetland soils under both anaerobic and aerobic conditions. Journal of Environmental Sciences, 2010, 22(8): 1232- 1238.
[63] Yun J L, Yu Z S, Li K, Zhang H X. Diversity, abundance and vertical distribution of methane-oxidizing bacteria (methanotrophs) in the sediments of the Xianghai wetland, Songnen Plain, Northeast China. Journal of Soils and Sediments, 2013, 13 (1):242- 252.
[64] Wasmund K, Kurtböke D I, Burns K A, Bourne D G. Microbial diversity in sediments associated with a shallow methane seep in the tropical Timor Sea of Australia reveals a novel aerobic methanotroph diversity. FEMS Microbiology Ecology, 2009, 68(2): 142- 151.
[65] Tavormina P L, Ussler W, Joye S B, Harrison B K, Orphan V J. Distributions of putative aerobic methanotrophs in diverse pelagic marine environments. The ISME Journal, 2010, 4(5): 700- 710.
[66] Durisch-Kaiser E, Klauser L, Wehrli B, Schubert C. Evidence of intense archaeal and bacterial methanotrophic activity in the Black Sea water column. Applied and Environmental Microbiology, 2005, 71(12): 8099- 8106.
[67] Kolb S, Knief C, Dunfield P F, Conrad R. Abundance and activity of uncultured methanotrophic bacteria involved in the consumption of atmospheric methane in two forest soils. Environmental Microbiology, 2005, 7(8): 1150- 1161.
[68] Horz H P, Rich V, Avrahami S, Bohannan B J M. Methane-oxidizing bacteria in a California upland grassland soil: Diversity and response to simulated global change. Applied and Environmental Microbiology, 2005, 71(5): 2642- 2652.
[69] Mohanty S R, Bodelier P L E, Floris V, Conrad R. Differential effects of nitrogenous fertilizers on methane-consuming microbes in rice field and forest soils. Applied and Environmental Microbiology, 2006, 72(2): 1346- 1354.
[70] Menyailo O V, Abraham W R, Conrad R. Tree species affect atmospheric CH4oxidation without altering community composition of soil methanotrophs. Soil Biology and Biochemistry, 2010, 42(1): 101- 107.
[71] Baani M, Liesack W L. Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found inMethylocystissp. Strain SC2. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(29): 10203- 10208.
[72] Dedysh S N, Khmelenina V N, Suzina N E, Trotsenko Y A, Semrau J D, Liesack W, Tiedje J M.Methylocapsaacidiphilagen. nov., sp nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium fromSphagnumbog. International Journal of Systematic and Evolutionary Microbiology, 2002, 52(1): 251- 261.
[73] Dunfield P F, Khmelenina V N, Suzina N E, Trotsenko Y A, Dedysh S N.Methylocellasilvestrissp. nov., a novel methanotroph isolated from an acidic forest cambisol. International Journal of Systematic and Evolutionary Microbiology, 2003, 53(5): 1231- 1239.
[74] Trotsenko Y A, Khmelenina V N. Biology of extremophilic and extremotolerant methanotrophs. Archives of Microbiology, 2002, 177(2): 123- 131.
[75] Heyer J, Berger U, Hardt M, Dunfield P F.Methylohalobiuscrimeensisgen. nov., sp. nov., a moderately halophilic, methanotrophic bacterium isolated from hypersaline lakes of Crimea. International Journal of Systematic and Evolutionary Microbiology, 2005, 55(5): 1817- 1826.
[76] Omel′Chenko M V, Vasil′Eva L V, Zavarzin G A, Savel′Eva N D, Lysenko A M, Mityushina L L, Khmelenina V N, Trotsenko Y A. A novel psychrophilic methanotroph of the genusMethylobacter. Microbiology, 1996, 65(3): 339- 343.
[77] Bodrossy L, Holmes E M, Holmes A J, Kovács K L, Murrell J C. Analysis of 16S rRNA and methane monooxygenase gene sequences reveals a novel group of thermotolerant and thermophilic methanotrophs,Methylocaldumgen. nov. Archives of Microbiology, 1997, 168(6): 493- 503.
[78] Bodrossy L, Kovács K L, McDonald I R, Murrell J C. A novel thermophilic methane-oxidising γ-Proteobacterium. FEMS Microbiology Letters, 1999, 170(2): 335- 341.
[79] Tsubota J, Eshinimaev B T, Khmelenina V N, Trotsenko Y A.Methylothermusthermalisgen. nov., sp. nov., a novel moderately thermophilic obligate methanotroph from a hot spring in Japan. International Journal of Systematic and Evolutionary Microbiology, 2005, 55(5): 1877- 1884.
[80] Kip N, Van Winden J F, Pan Y, Bodrossy L, Reichart G J, Smolders A J P, Jetten M S M, Damsté J S S, Op den Camp H J M. Global prevalence of methane oxidation by symbiotic bacteria in peat-moss ecosystems. Nature Geoscience, 2010, 3(9): 617- 621.
[81] Chen Y, Murrell J C. Geomicrobiology: Methanotrophs in moss. Nature Geoscience, 2010, 3(9): 595- 596.
[82] Kip N, Ouyang W J, van Winden J, Raghoebarsing A, van Niftrik L, Pol A, Pan Y, Bodrossy L, van Donselaar E G, Reichart G J, Jetten M S M, Damste J S S, and den Camp H J M O. Detection, Isolation, and characterization of acidophilic methanotrophs fromSphagnumMosses. Applied and Environmental Microbiology, 2011, 77(16): 5643- 5654.
[83] Kip N, Fritz C, Langelaan E S, Pan Y, Bodrossy L, Pancotto V, Jetten M S M, Smolders A J P, Op den Camp H J M. Methanotrophic activity and diversity in differentSphagnummagellanicumdominated habitats in the southernmost peat bogs of Patagonia. Biogeosciences, 2012, 9(1): 47- 55.
[84] Liebner S, Zeyer J, Wagner D, Schubert C, Pfeiffer E M, Knoblauch C. Methane oxidation associated with submerged brown mosses reduces methane emissions from Siberian polygonal tundra. Journal of Ecology, 2011, 99(4): 914- 922.
[85] Knief C, Dunfield P F. Response and adaptation of different methanotrophic bacteria to low methane mixing ratios. Environmental Microbiology, 2005, 7(9): 1307- 1317.
[86] Bodelier P L E, Laanbroek H J. Nitrogen as a regulatory factor of methane oxidation in soils and sediments. FEMS Microbiology Ecology, 2004, 47(3): 265- 277.
[87] Mosier A, Schimel D, Valentine D, Bronson K, Parton W. Methane and nitrous oxide fluxes in native, fertilized and cultivated grasslands. Nature, 1991, 350(6316): 330- 332.
[88] Bodelier P L E, Roslev P, Henckel T, Frenzel P. Stimulation by ammonium-based fertilizers of methane oxidation in soil around rice roots. Nature, 2000, 403(6768): 421- 424.
[89] Murase J, Frenzel P. A methane-driven microbial food web in a wetland rice soil. Environmental Microbiology, 2007, 9(12): 3025- 3034.
[90] Murase J, Hordijk K, Tayasu I, Bodelier P L E. Strain-specific incorporation of methanotrophic biomass into eukaryotic grazers in a rice field soil revealed by PLFA-SIP. FEMS Microbiology Ecology, 2011, 75(2): 284- 290.
[91] Moon L E, Lee S Y, Lee S H, Ryu H W, Cho K S. Earthworm cast as a promising filter bed material and its methanotrophic contribution to methane removal. Journal of Hazardous Materials, 2010, 176(1/3): 131- 138.
[92] Kim T G, Moon K E, Lee E H, Choi S A, Cho K S. Assessing effects of earthworm cast on methanotrophic community in a soil biocover by concurrent use of microarray and quantitative real-time PCR. Applied Soil Ecology, 2011, 50: 52- 55.
[93] Zhou X Q, Wang Y F, Huang X Z, Hao Y B, Tian J Q, Wang J Z. Effects of grazing by sheep on the structure of methane-oxidizing bacterial community of steppe soil. Soil Biology and Biochemistry, 2008, 40(1): 258- 261.
[94] Zheng Y, Yang W, Sun X, Wang S P, Rui Y C, Luo C Y, Guo L D. Methanotrophic community structure and activity under warming and grazing of alpine meadow on the Tibetan Plateau. Applied Microbiology and Biotechnology, 2012, 93(5): 2193- 2203.
参考文献:
[29] 韩冰, 苏涛, 李信, 邢新会. 甲烷氧化菌及甲烷单加氧酶的研究进展. 生物工程学报, 2008, 24(9): 1511- 1519.
Ecologyofaerobicmethaneoxidizingbacteria(methanotrophs)
YUN Juanli, WANG Yanfen*, ZHANG Hongxun
GraduateUniversityofChineseAcademyofSciences,Beijing100049,China
Aerobic methane oxidizing bacteria (methanotrophs) are a fascinating group of bacteria that have the unique ability to grow on methane as their sole carbon and energy source. They appear to be widespread in nature and have been isolated from a number of different environments. There are now 14 recognized genera of methanotrophs belong to two phyla, Proteobacteria and thermoacidiphilic Verrucomicrobia. The former was well studied and separated into two classes, TypeⅠ and TypeⅡ methanotrophs, which belong to Alpha and Gamma Proteobacteria. Extremely thermophilic, acidophilic methanotrophs from the phylum Verrucomicrobia have been isolated, thus expanding both the taxonomic diversity and physiological range of aerobic methanotrophy.
The discovery of the facultative methanotrophMethylocellasilvestrishas changed the view that methanotrophs were obligate organism. They can cooxidize a considerable number of organic compounds and also have considerable potential in biotechnology. A wide variety of methanotrophic symbionts in and on the mosses were recently detected, and showing the global prevalence of this symbiosis. Traditional way used cultivation to enrichment or isolation to study methanotrophs in the environment. Molecular ecology techniques applied in the last few decades have greatly expanded our knowledge of methanotroph ecology. The most obvious marker for detecting methanotrophs in various environments is the 16S rRNA gene, due to the large database of sequences available. Primers and probes targeting different genera or species have been designed and used extensively in combination with polymerase chain reaction (PCR) based clone library analysis, denaturing gradient gel electrophoresis (DGGE) analysis, and fluorescent in situ hybridization (FISH) analysis. Several functional genes have also been used for the detection of methanotrophs in environmental samples, includingpmoA(encoding the key subunits of particulate methane monooxygenase),mmoX(encoding the key subunits of soluble methane monooxygenase),mxaF(encoding the key subunits of methanol dehydrogenase),nifH(encoding the dinitrogenase reductase), and genes involved in C1 transfer pathways. To understand the active community of methanotrophs in the environment, stable isotope probing (SIP) techniques have been developed, including DNA-SIP, RNA-SIP, mRNA-SIP, and phospholipid fatty acid (PLFA)-SIP. SIP has also been combined with metagenomics to discover novel methanotrophs. Other very powerful molecular techniques have been developed in the last few years, including microautoradiography (MAR)-FISH, isotope array, Raman-FISH, nano-secondary ion mass spectrometry (NanoSIMS), and microfluidic digital PCR, these techniques can now be used in the analyses of methanotrophs. Both cultivation and cultivation independent molecular methods have been used intensively in last few decades to study the diversity, distribution, and abundance in environments of methanotrophs, such as soils, freshwater, marine sediments, acid peat bogs, hot springs, seawater and extreme environments. In the microcosm of soil, the growth and diversity of methanotrophs are also influenced by several environmental factors. This review highlights recent progress in the research of the taxonomy, of the discovery of novel aerobic methanotrophs, of the biochemistry, of the molecular techniques and the environment impacts on methanotrophs, we also emphasize deficiencies and issues need to be solved in future studies. This review will provide theoretical foundation for future methanotrophic ecology study and explain the key role methanotrophs play in carbon cycle.
methanotrophs; microbial ecology; taxonomy; diversity; carbon-cycle
国家自然科学基金资助项目(41271277/D010504)
2012- 07- 17;
2013- 04- 24
*通讯作者Corresponding author.E-mail: yfwang@ucas.ac.cn
10.5846/stxb201207171013
贠娟莉,王艳芬,张洪勋.好氧甲烷氧化菌生态学研究进展.生态学报,2013,33(21):6774- 6785.
Yun J L, Wang Y F, Zhang H X.Ecology of aerobic methane oxidizing bacteria (methanotrophs).Acta Ecologica Sinica,2013,33(21):6774- 6785.
