Cloning and expression analysis of a long type peptidoglycan recognition protein (PGRP-L) from Xenopus tropicalis
2011-12-25QIZhiTaoZHANGQiHuanWANGZiShengWANGAiMinHUANGBeiCHANGMingXianNIEPin
QI Zhi-Tao, ZHANG Qi-Huan, WANG Zi-Sheng, WANG Ai-Min, HUANG Bei, CHANG Ming-Xian, NIE Pin
(1. Department of Ocean Technology, Key Laboratory of Aquaculture and Ecology of Coastal Pools of Jiangsu Province, Yancheng Institute of Technology, Yancheng 224051, China; 2. Central Laboratory of Biology, Chemical and Biological Engineering College, Yancheng Institute of Technology, Yancheng 224051, China; 3. State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, the Chinese Academy of Sciences, Wuhan 430072, China)
Cloning and expression analysis of a long type peptidoglycan recognition protein (PGRP-L) fromXenopus tropicalis
QI Zhi-Tao1,2,3,*, ZHANG Qi-Huan2, WANG Zi-Sheng1, WANG Ai-Min1, HUANG Bei3, CHANG Ming-Xian3, NIE Pin3
(1.Department of Ocean Technology,Key Laboratory of Aquaculture and Ecology of Coastal Pools of Jiangsu Province,Yancheng Institute of Technology,Yancheng224051,China; 2.Central Laboratory of Biology,Chemical and Biological Engineering College,Yancheng Institute of Technology,Yancheng224051,China; 3.State Key Laboratory of Freshwater Ecology and Biotechnology,Institute of Hydrobiology,the Chinese Academy of Sciences,Wuhan430072,China)
Peptidoglycan recognition proteins (PGRPs) are a family of pattern recognition receptors (PRRs) of the immune system, which bind and hydrolyze bacterial peptidoglycan. Here, a long type PGRP (PGRP-L) was first cloned in the lower vertebrate speciesXenopus tropicalis(Xt). The XtPGRP-L possessed a conserved genomic structure with five exons and four introns. The alignment and phylogenetic analysis indicated that XtPGRP-L might be a type of amidase-like PGRP. The 3-D model showed that XtPGRP-L possessed a conserved structure compared with the Drosophila PGRP-Lb. During embryonic development, XtPGRP-L was not expressed until the 72 h tadpole stage. In adult tissues, it was strongly expressed in the liver, lung, intestine, and stomach. Furthermore, after LPS stimulation, the expression of XtPGRP-L was up-regulated significantly in the liver, intestine and spleen, indicating that XtPGRP-L may play an important role in the innate immunity ofXenopus tropicalis.
Peptidoglycan recognition protein; Gene clone; Expression analysis;Xenopus tropicalis
Peptidoglycan recognition proteins (PGRPs) are a group of receptors that recognize conserved pathogenassociated molecular patterns (PAMPs) to initiate host immune responses. They were first identified in thehemolymph and cuticle of the silkworm (Bombyx mori), and were shown to bind peptidoglycan (PGN) and activate the prophenoloxidase cascade (Yoshida et al, 1996). Members of the PGRP family have been characterized in insects, teleosts, and mammals and are evolutionarily conserved. Twelve PGRP genes have been reported inDrosophila melanogaster, which are classified into two classes based on length: the short class (PGRP-S) including PGRP-SA, SB1, SB2, SC1A, SC1B, SC2, and SD; and the long class (PGRP-L) including PGRP-LA, LB, LC, LD, and LE. Insect PGRPs have diverse functions and roles such as cell activation and phagocytosis (Werner et al, 2000). In teleost, multiple PGRP genes have been discovered (Chang et al, 2007), some of which exhibit peptidoglycan-lytic amidase activity and broad-spectrum bactericidal activity (Li et al, 2007), and are involved in various pathways (Chang&Nie, 2008; Chang et al, 2009; Wang&Lai, 2010). Unlike insects and fish, only four PGRPs have been identified in mammals (Liu et al, 2000, 2001): PGRP-S (short), PGRP-IA (I for intermediate), PGRP-IB (intermediate), and PGRP-L (long), which also have amidase and antibacterial activities (Li et al, 2007). However, PGRPs have not been reported in other groups of vertebrates.
Using the available genome database of the clawed frog (Xenopus tropicalis), it is possible to identify PGRPs from this amphibian model. In this study, a single long type PGRP was first identified fromX. tropicalis, with its genomic structure and expression pattern in ontogeny and adult in response to LPS stimulation also investigated.
1 Material and Methods
1.1 Database mining and cloning for Xenopus PGRPL (XtPGRP-L)
To search for possible PGRP homologues, the reported PGRP protein sequences fromDrosophilaand mammals were used to perform tBLASTp search in theXenopusgenome database (http://www.ensembl.org). The obtained DNA sequence was then analyzed for predicted transcripts using GenScan at the Massachusetts Institute of Technology (http://genes.mit.edu/GENSCAN. html). To determine the correct reading frame of each exon, the BLAST program was used to maximize the inframe translated sequence. A PGRP-L was identified in the database.
To obtain the full length of the identified XtPGRPL gene, total RNA was extracted fromXenopusliver using Trizol reagent (Invitrogen, USA). Approximately 500 ng total RNA was used as the template for the synthesis of first strand cDNA by reverse transcriptase using superscript II reverse transcription system (Invitrogen, USA). The full-length cDNA sequence of XtPGRP-L was amplified by random amplification of cDNA ends (RACE). The 5′ end region of the PGRP-L was amplified by nested PCR with two primer pairs of UPM/PGRP-LRout (1stround PCR) and UPM/PGRPLRin (2ndround). The gene specific primers used for 3′RACE PCR were UPM/PGRP-LFout (1stround PCR) and UPM/PGRP-LFin (2ndround). The PCR was carried out as per the following: in 25 μL solution containing 125 μmol of each dNTP, 0.2 μmol of each primer, 2.5 μL 10 × Taq buffer, 12 U Ex Taq polymerase, 18.1 μL sterile H2O and 1 μL cDNA template, according to the standard protocol, and under the following conditions: an initial cycle of denaturation step at 94 °C for 5 min, followed by 8 cycles of amplification at 94 °C for 30 s, 64 °C for 30 s, 72 °C for 1 min, and 30 cycles at 94 °C for 30 s, 62 °C for 30 s, 72 °C for 1 min, and a final extension step at 72 °C for 10 min. A total of 10 μL of product was size-fractioned by 1.5% (W/V) agarose gel electrophoresis and stained with ethidium bromide. The desired PCR products were isolated using the Omega agarose purification kit, and cloned into pMD18-T vectors (TaKaRa) according to manufacturer’s instructions, before being transformed intoEscherichia coliDH5α competent cells and plated on selective agar plates containing 100 μg/mL of ampicillin. Positive colonies were grown overnight, and then determined using specific primers under the same cycle of PCR, before being sequenced using dideoxy chain-termination on an automatic DNA sequencer (ABI Applied Biosystems Mode 377). All primers used are listed in Tab.1.

Tab. 1 Primer sequences
1.2 Sequence analysis of XtPGRP-L
The XtPGRP-L amino acid sequence was deduced using the Expasy translation tool (http://ca.expasy.org/ tools). The alignment between the amino acid sequence of XtPGRP-L and other known PGRPs was analyzed using the CLUSTAL W program (version 1.83) (Thompson et al, 1994) and decorated using GeneDoc (http://www. nrbsc.org/gfx/genedoc/index.html). Identities between PGRP sequences were determined using the Megalign program within DNASTAR. A phylogenetic tree was constructed using the neighbor-joining (N-J) method within the Mega4.0 software program (http://www. megasoftware.net/mega.html) and all sequences were obtained from the NCBI database and are listed in Tab. 2. The 2-D structure of XtPGRP-L was predicted at PSIPREDView (http://bioinf.cs.ucl.ac.uk/psipred/psiform. html) (Jones, 1999; McGuffin et al, 2000). The 3-D structure of XtPGRP-L was constructed in the SWISSMODEL protein modeling server (http://expasy.org/ swissmod/SWISS-MODEL. html). A suitable structural template for XtPGRP-L,Drosophila melanogaster(Dm) PGRP-Lb (PDB file code: 1OHT-A), was identified by a BLAST search as implemented in the SWISS-MODEL protein modeling server (http://expasy.org/swissmod/ SWISS-MODEL.html) (Arnold et al, 2006). The automatic sequence alignment obtained was used for homology modeling in SWISS-MODEL (Schwede et al, 2003), and the resultant theoretical model was displayed and analyzed with Swiss-PDB Viewer. The model picture was drawn using the Rasmol program (http:// www.umass.edu/microbio/rasmol/Rasmol).

Tab. 2 Peptidoglycan recognition protein sequences used for phylogenetic tree construction and multiple sequence alignment
1.3 Expression of XtPGRP-L mRNA in developing embryos and in different organs
To characterize the expression of XtPGRP-L in relation toXenopusdevelopment and in different organs, developing embryos were collected at 3, 6, 15, 48, 72, 144, 216, and 360 h post-fertilization (hpf) for total RNA isolation (Dale & Slack, 1987). The RT-PCR was performed according to Liang et al. (2006), and the gene specific primers for RT-PCR were designed to the XtPGRP-L fragment and loading control primers were designed according toXenopusβ-actin. The primers used are listed in Tab.1.
To reveal the expression in different organs of adultXenopus, heart, liver, lung, kidney, intestine, stomach and spleen were collected for total RNA preparation and quantitative -PCR (Qi & Nie, 2008). All primers used are listed in Tab.1.
1.4 Preparation of polyclonal antibody against XtPGRP-L and Western blotting analysis
The 1 037 bp expression fragment of XtPGRP-L was obtained by one pair of primers containingKpnI andHind III digestion sites (Ex-PGRPLF/R; Tab. 1). The PCR amplifications were performed at one cycle of 94 °C for 5 min, 33 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 60 s, with a final extension step of 72 °C for 10 min. Purified fragments were digested withKpnI andHind III, then ligated into the pQE-40 expression vector for recombinant protein construction, and transformed intoEscherichia coliM15 competent cells (TAKARA) for protein expression. Fusion protein was expressed by isopropyl-β-D-thiogalactopyranoside (IPTG) induction, and analyzed by SDS polyacrylamide gel (SDS-PAGE). Compared with the un-induced sample, a predominant extra protein band with molecular weight of about 58 000 was observed in the IPTG induced samples. The recombinant protein was purified by affinity chromatography in a column of Ni2+-charged resin (Novagen) according to manufacturer’s instructions. To prepare the antibody, the purified recombinant protein was used to immunize rabbit. Briefly, the rabbit was first injected with 800 mg purified protein. Every two weeks, the rabbit was then immunized twice with half the initial amount of proteins. Two weeks after the last injection, the rabbit was bled and serum was collected and stored at -80°C.
To study the expression of XtPGRP-L at the protein level, heart, liver, spleen, lung, kidney, intestine and stomach were dissected and homogenized in buffer containing 1% SDS, 5% β-mercaptoethanol, 10% glycerol, 65 mmol Tris-HCl, pH 6.8 and 1 mmol PMSF. The suspension was vortexed and centrifuged at 10 000gfor 10 min, and the supernatant was retained. Protein concentrations were determined using Bradford protein assay (Biorad). Each sample, equivalent to 10 mg total protein, was run on 12% SDS-PAGE and subsequently transferred to PVDF membrane (Millipore) by standard procedures. The blotting membrane was blocked overnight at 4 °C with TBS containing skimmed milk and 0.05% Tween 20, before being incubated for 2 h with rabbit antiserum (1:300). Subsequently, the membrane was further incubated for 1.5 h with Alkaline Phosphatase goat Anti-Rabbit IgG (Boster, China). After washing, the specific binding to recombinant protein was detected by NBT/BCIP (SABC).
1.5 Real-time quantitative PCR analysis of XtPGRPL expression
Quantitative real-time PCR was performed to determine the expression of XtPGRP-L in normal frogs and those injected with LPS. Two groups ofXenopus, each containing three frogs, were injected intraperitoneally with phosphate-buffered saline (PBS, pH7.2) for the control and LPS (150 μg/100g body weight) for the experiment, respectively. Twenty-four hours later,Xenopuswere euthanized and tissues were collected for total RNA extraction and cDNA synthesis. The housekeeping gene β-actin amplified by actin-F/actin-R was used as the control. The primers used for real-time quantitative PCR analysis are listed in Tab. 1.
Quantitative real-time PCR was conducted on Chromo 4TMContinuous Fluorescence Detection from MJ Research. Amplifications were carried out at 20 µL volume, containing 1 µL of cDNA template, 10 µL of SYBR green Mix (Toyobo, Japan), 0.3 µL of each primer and 8.4 µL of ddH2O. The PCR amplification was performed in triplicate wells, using the following conditions: 3 min at 95 °C, followed by 40 cycles consisting of 30 s at 94 °C, 25 s at 58 °C and 25 s at 72 °C. Melting curve analysis of the amplification products was performed at the end of each PCR reaction to confirm that a single PCR product was detected.
Expression values were calculated using the relative standard curve method provided by the ABI Sequence Detection System software. Data analysis was done according to Purcell et al (2004). Statistical analyses (t-tests) were performed using Microcal origin 6.0 withP≤0.05 as the significance level.
2 Results
2.1 Sequence characteristics and phylogenetic analysis
A genomic scaffold_149 of 2.34 Mb in size was identified fromXenopusgenome database. About 129 transcripts were predicted using the FGENESH program. Using Netblast analysis, the transcript that shared high identity with PGRP-L was verified. The XtPGRP-L cDNA sequence contained a 40 bp 5′-untranslated region (UTR), a 1 491 bp open reading frame, and a 150 bp 3′-UTR with a discernable poly (A) addition signal (AATAAA) (GenBank accession no. EF095491). The genome structure is conserved inXenopus, zebrafish, mouse and human PGRP-L, which all contain five exons and four introns of similar size. Typical intron splice motifs were observed at the 5′(gt) and 3′(ag) ends of each intron. Putative XtPGRP-L possessed 497 amino acids, with a calculated molecular mass of 55 000 and an isoelectric point of 8.11. Analysis by SignalP showed that XtPGRP-L contained an 18-aa signal peptide (MAKVLLILLCTLCAMAVA). Similar to human, mouse and zebrafish PGRP-L, the XtPGRP-L also possessed a transmembrane domain (Fig.1).
Comparing the PGRP-L genes with PGRP-L from other vertebrates showed that all PGRP-L genes contained a conserved PGRP domain of approximately 160 amino acids (Fig.2). The identity between XtPGRPL and other reported PGRP-L ranged from 31.3 % -66.7%. Further analysis showed that this domain can be subdivided into three PGRP domains (I, II and III). The three domains in XtPGRP-L had 8.3% to 51.2% identities, while domains II and III had the highest
(51.2%). The three PGRP domains in XtPGRP-L shared 52.6%, 87.5% and 78.0% identities with human counterparts, respectively. Although the identity between XtPGRP-L and T7 lysozyme (27.4% identity) was low, the positions of the amino acids needed for enzyme activity in T7 lysozyme, such as His-17, Gly40, Trp41, Tyr46, Arg60, His122 and Cys-130, were conserved (Fig. 2). The 2-D structure of XtPGRP-L predicted by PSIPREDView showed that XtPGRP-L was an αhelix/β-strand mixed protein containing four α-helices and seven β-strands (Fig. 3a). The 3-D structure of XtPGRP-L was predicted by comparative modeling usingDrosophila melanogasterPGRP-Lb (PDB file code: 1OHT-A) as a template. The model obtained from SWISS-MODEL was in accordance with the 2-D structure and showed that XtPGRP-L contained four αhelices and seven β-sheets. Compared with the Dm PGRP-LB (GenBank accession no. NP_731576), XtPGRP-L possessed a conserved structure: β2-β3-α1-β4-β5-β6- α2-β7-α3. But, the XtPGRP-L has another αhelix (α*) and β-sheet (β*) (Fig. 3b).

Fig. 1 Comparing the XtPGRP-L protein and gene structure with other vertebrates’ long type PGRPs

Fig. 2 Multiple alignment of XtPGRP-L with other PGRPs

Fig. 3 The predicted secondary structure and homology modeling of XtPGRP-L
To further analyze the evolutionary relationship between XtPGRP-L and other PGRPs, an unrooted phylogenetic tree was constructed using the Neighbor-Joining methods. The XtPGRP-L was grouped closely with other vertebrate PGRP-L with high bootstrap values (about 99%). Surprisingly, the vertebrate PGRP-L was not clustered with the long type PGRP fromDrosophila melanogaster(Fig.4). A previous study showed that PGRP-S may be the oldest PGRP type and evolved PGRP-L much later (Dziarski, 2004; Qi et al, 2010). Thus, theDrosophila melanogasterand vertebrates PGRP-L may evolve from a common PGRP-S ancestor gene and expanded by gene duplication.
2.2 Expression of XtPGRP-L gene in developing embryos and normal adults and in response to LPS stimulation
Using the XtPGRP-L specific primers, XtPGRP-L mRNA was not detected in early development stages (3 -48 h) until 72 h (Fig. 5a). In normal adultXenopus, XtPGRP-L mRNA was detected in liver, lung, intestine, stomach, heart, spleen and kidney (Fig. 5b, d). The XtPGRP-L was detected at the protein level in all organs examined by Western blotting. As shown in Fig. 5c, an immunoreactive band at about 55 000 was observed.
The XtPGRP-L expression pattern after LPS stimulation was examined using quantitative real-time PCR. The XtPGRP-L expression was obviously upregulated in the heart, liver, spleen and intestine after LPS treatment, with of 3-, 26- 7- and 33- fold increases, respectively. However, significant changes were not detected in the lung, kidney, and stomach (Fig. 5e).
3 Discussion
The present work described for the first time a long type PGRP fromX. tropicalisby analyzing its genome database. The genomic structure of this PGRP and its expression pattern were analyzed.
The XtPGRP-L was 497-aa in length and contained a conserved genome structure, as in human, mouse and zebrafish. Furthermore, the XtPGRP-L also contained a highly conserved PGRP domain and shared high sequence identities with mammalian long type PGRP (eg. 66.7% withMus musculus; 65.3% withHomo sapiens). The phylogenetic tree showed that XtPGRP-L grouped closely with PGRP-L from other vertebrates. These results confirmed that the cloned gene belonged to the long type PGRP. The predicted 3-D structure for the XtPGRP-L showed that the mixed β-sheet of six strands and three α-helices that formed the protein core was conserved in evolution. Furthermore, the Cys, Tyr and Thr residues located in β3-α1, α1-β4 and β7-α3 loops, respectively, are essential for PGN recognition (Reiser et al, 2004).

Fig. 4 An unrooted phylogenetic tree constructed by the neighbor joining from amino acid sequences of PGRPs together with the XtPGRP-L gene
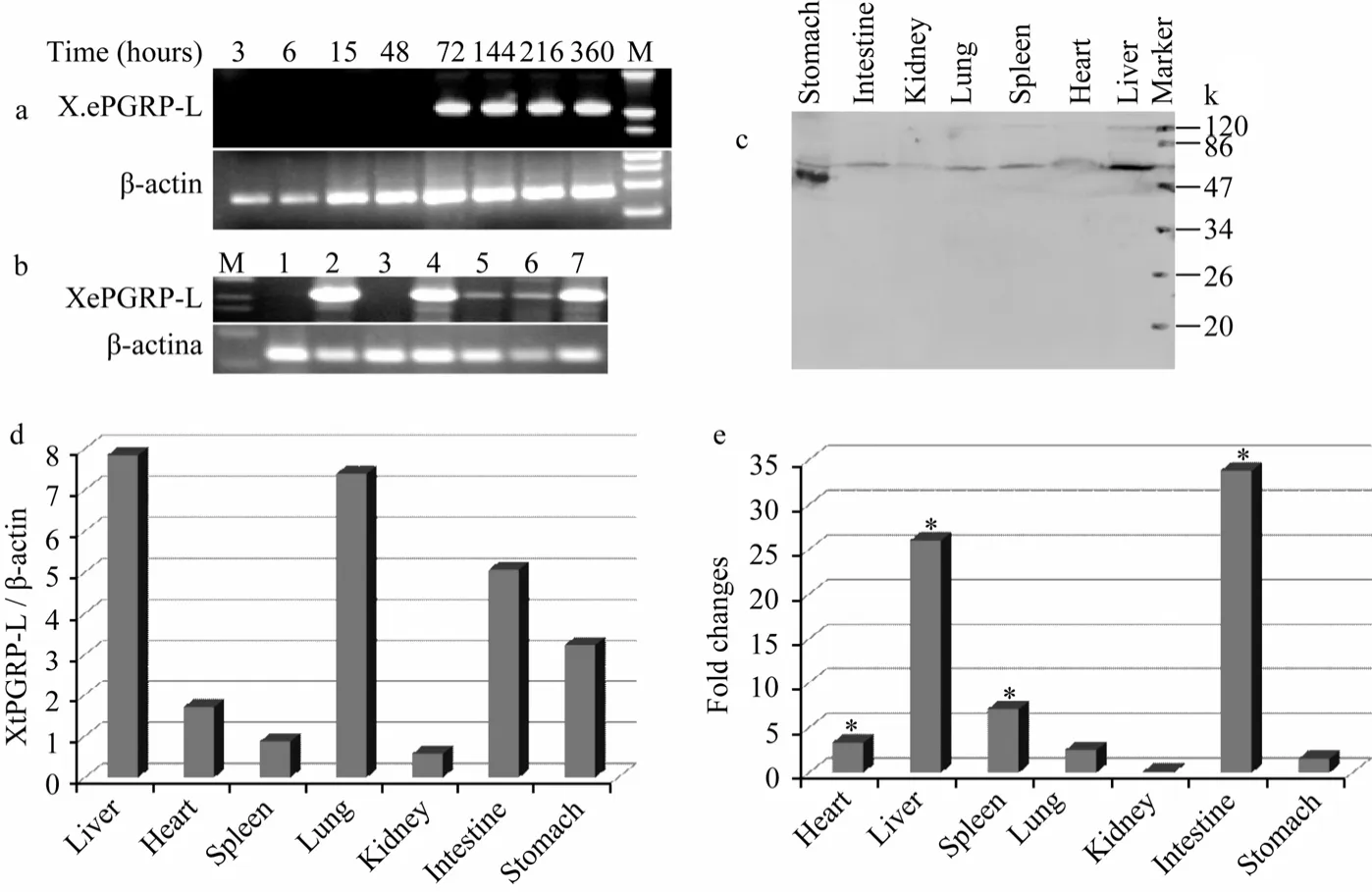
Fig. 5 Expression pattern of the XtPGRP-L gene in developing embryos, tissues of normal Xenopus and LPS stimulated Xenopus
In T7 lysozyme, His-17, His-122, and Cys-130 are Zn2+binding ligands, and Tyr-46 and Lys-128 are needed for enzymatic activity. Site directed mutagenesis of T7 lysozyme has shown that activity is retained when Lys-128 is replaced by Thr (Cheng et al, 1994). Divalent cations are required for the amidase activity of human PGRP-L, and Zn2+fully restores its amidase activity. Among of four mammalian PGRPs, only PGRP-L has all amino acids needed for the amidase activity of T7 lysozyme conserved. In human PGRP-L, the Zn2+binding amino acids and Cys419 are required for the amidase activity, whereas the three other amino acids, needed for the activity of T7 amidase, are not required (Wang et al, 2003). The alignment of the XtPGRP-L with T7 lysozyme and other PGRPs showed that residues needed for T7 lysozyme activity, were found in XtPGRP-L and other PGRPs proved to be amidase (Fig.3) (Steiner, 2004). This analysis suggests that XtPGRP-L might also be an amidase.
The expression pattern of PGRPs is different from insect to mammalian. In insects, many PGRPs are expressed in immune competent organs, consistent with their role in insect immunity (Dimopoulos et al, 2002). Long insect PGRPs are mainly expressed in hemocytes. Human PGRPs have highly differential expression in various organs and tissues (Dziarski, 2004). In the present study,XenopusPGRP-L mRNA, similar to human PGRP-L, was expressed in a wide range of tissues including liver, lung, kidney, stomach, and intestine, although there may be quantitative differences. Furthermore, after LPS stimulation, the expression of XtPGRP-L was significantly up-regulated in several tissues, which suggests an important role for XtPGRP-L in the fight against bacterial infections in adultXenopus. Furthermore, the XtPGRP-L expressed in the developing embryo armed the embryo to defend against bacterial infections before the development of adaptive immunity.
Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling [J].Bioinformatics, 22: 195-201.
Chang MX, Nie P, Wei LL. 2007. Short and long peptidoglycan recognition proteins (PGRPs) in zebrafish, with findings of multiple PGRP homologs in teleost fish [J].Mol Immunol, 44: 3005-3023.
Chang MX, Nie P. 2008. RNAi suppression of zebrafish peptidoglycan recognition protien 6 (zfPGRP6) mediated differentially expressed genes involved in Toll-like receptor signaling pathway and caused increased susceptibility toFlavobacterium columnare[J].Vet Immunol Immunopathol, 124: 295-301.
Chang MX, Wang YP, Nie P. 2009. Zebrafish peptidoglycan recognition protein SC (zfPGRP-SC) mediates multiple intracellular signaling pathways [J].Fish Shellfish Immunol, 26: 264-274.
Cheng X, Zhang X, Pflugrath JW, Studier FW. 1994. The structure of bacteriophage T7 lysozyme, a zinc amidase and an inhibitor of T7 RNA polymerase [J].Proc Natl Acad Sci USA, 91: 4034-4038.
Dale L, Slack JM. 1987. Fate map for the 32-cell stage ofXenopus laevis[J].Development, 99: 527-551.
Dimopoulos G, Christophides GK, Meister S, Schultz J, White KP, Barillas-Mury C, Kafatos FC. 2002. Genome expression analysis of Anopheles gambiae: Responses to injury, bacterial challenge, and malaria infection [J].Proc Natl Acad Sci USA, 99: 8814-8819.
Dziarski R. 2004. Peptidoglycan recognition proteins (PGRPs) [J].Mol Immunol, 40: 877-886.
Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices [J].J Mol Biol, 292:195-202.
Li XN, Wang SY, Qi J, Echtenkamp SF, Chatterjee R, Wang M, Boons GJ, Dziarski R, Gupta D. 2007. Zebrafish peptidoglycan recognition proteins are bactericidal amidases essential for defense against bacterial infections [J].Immunity, 27: 518–529.
Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA. 2006. Interleukin 22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides [J].J Exp Med, 203: 2271–2279.
Liu C, Gelius E, Liu G, Steiner H, ziarski R. 2000. Mammalian peptidoglycan recognition protein binds peptidoglycan with high affinity, is expressed in neutrophils, and inhibits bacterial growth [J].J Biol Chem, 275: 24490-24499.
Liu C, Xu Z, Gupta D, Dziarski R. 2001. Peptidoglycan recognition proteins: A novel family of four human innate immunity pattern recognition molecules [J].J Biol Chem, 276: 34686–34694.
McGuffin L, Bryson K, Jones DT. 2000. The PSIPRED protein structure prediction server [J].Bioinformatics, 16: 404-405.
Purcell MK, Kurath G, Garver KA, Herwig RP, Winton JR. 2004. Quantitative expression profiling of immune response genes in rainbow trout following infectious haematopoietic necrosis virus (IHNV) infection or DNA vaccination [J].Fish Shellfish Immunol, 17: 262-447.
Qi ZT, Nie P. 2008. Comparative study and expression analysis of the interferon gamma gene locus cytokines inXenopus tropicalis[J].Immunogenetics, 60:699-710.
Qi ZT, Gao Q, Huang B, Chang MX, Nie P. 2010. Cloning and identification of a short type Peptidoglycan recognition protein inXenopus tropicalis[J].Acta Hyrobiol Sin, 34:922-926. (in Chinese)
Reiser JB, Teyton L, Wilson IA. 2004. Crystal structure of the Drosophila peptidoglycan recognition protein (PGRP)-SA at 1.56 A resolution [J].J Mol Biol, 340: 909-917.
Schwede T, Kopp J, Guex N, Peitsch MC. 2003. SWISS-MODEL: An automated protein homology-modeling server [J].Nucleic Acids Res, 31: 3381-3385.
Steiner H. 2004. Peptidoglycan recognition proteins: on and off switches for innate immunity [J].Immunol Rev, 198: 83-96.
Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice [J].Nucleic Acids Res, 22: 4673-4680.
Wang YP, Lai R. 2010. Insect antimicrobial peptides: structures, properties and gene regulation [J].Zool Res, 31: 27-34. (in Chinese)
Wang ZM, Li X, Cocklin RR, Wang M, Fukase K, Inamura S, Kusumoto S, Gupta D, Dziarski R. 2003. Human peptidoglycan recognition protein-L is anN-acetylmuramoyl-L-alanine amidase [J].J Biol Chem, 278(49): 49044-49052.
Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. 2000. A family of peptidoglycan recognition proteins in the fruit flyDrosophila melanogaster[J].Proc Natl Acad Sci USA, 97: 13772-13777.
Yoshida H, Kinoshita K, Ashida M. 1996. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm,Bombyx mori[J].J Biol Chem, 271: 13854-13860.
非洲爪蟾中一种长型肽聚糖识别蛋白的克隆与表达分析
齐志涛1,2,3,*, 张启焕2, 王资生1, 王爱民1, 黄 贝3, 昌鸣先3, 聂 品3
(1. 盐城工学院 海洋技术系,江苏省沿海池塘养殖生态重点实验室, 江苏 盐城 224051; 2. 盐城工学院 化学与生物工程学院生物实验中心, 江苏 盐城 224051; 3. 中国科学院水生生物研究所 淡水生态与生物技术国家重点实验室, 湖北 武汉 430070)
肽聚糖识别蛋白(peptidoglycan recognition proteins, PGRPs)是固有免疫系统中一类重要的模式识别受体。该文首次从两栖类模式生物-非洲爪蟾 (Xenopus tropicalis)中克隆得到了一个长型 PGRP(XtPGRP-L)基因。XtPGRP-L具有5个外显子和4个内含子的基因组结构, 该结构在进化的过程中比较保守。序列比对与系统进化分析显示XtPGRP-L具有保守的酰胺酶活性位点。蛋白质建模显示XtPGRP-L拥有保守的3-D结构。实时定量PCR检测显示, XtPGRP-L在非洲爪蟾胚胎早期不表达, 到72 h蝌蚪期开始表达。在成体的肝脏、肺、肠和胃高表达。同时, 在LPS刺激后, XtPGRP-L在肝脏、肠和胃中呈明显上调表达。结果表明,XtPGRP-L在非洲爪蟾固有免疫系统中可能具有重要的作用。
肽聚糖识别蛋白; 基因克隆; 表达分析; 非洲爪蟾
Q959.53;Q786;R392.1
A
0254-5853-(2011)04-0371-08
2011-03-15;接受日期:2011-06-27
10.3724/SP.J.1141.2011.04371
date: 2011-03-15; Accepted date: 2011-06-27
s: This study was financially supported by the Project from the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (10KJB240001), the Foundation for Talent Recruitment of Yancheng Institute of Technology (XKR2011007), and the National Natural Science Foundation of China (30830083)*
(通信作者), E-mail: Tel: 0515-88298281, E-mail address: qizhitao@ycit.edu.cn
猜你喜欢
杂志排行
Zoological Research的其它文章
- 悬尾应激对小鼠空间记忆及其反转学习的损伤效应
- Visual modeling reveals cryptic aspect in egg mimicry of Himalayan Cuckoo (Cuculus saturatus) on its host Blyth’s Leaf Warbler (Phylloscopus reguloides)
- Afferent and efferent pathways in the visual system of the freshwater snail Planorbarius corneus
- Notch signaling dependent differentiation of cholangiocyte-like cells from rhesus monkey embryonic stem cells
- Metabolism and thermoregulation between Mrs Hume’s Pheasant (Syrmaticus humiae) and Elliot’s Pheasant (S. ellioti)
- Localization of stationary pronuclei during conjugation of Paramecium as indicated by immunofluorescence staining
