反刍家畜朊蛋白基因(PRNP)多态性及其遗传效应研究进展
2017-03-08魏振宇张少丽党瑞华雷初朝蓝贤勇
李 洁,魏振宇,彭 坤,张少丽,党瑞华,雷初朝,陈 宏,蓝贤勇
(1.西北农林科技大学动物科技学院,陕西 杨凌 712100;2. 西北农林科技大学创新实验学院,陕西 杨凌 712100)
朊蛋白基因(PRNP)不仅与动物传染性海绵样脑病(TSEs)密切相关,还与反刍家畜的表型性状密切相关。已有研究表明:牛、水牛、牦牛、绵羊、山羊等反刍家畜PRNP基因具有丰富的多态性,同时,这些遗传变异位点在这些物种中又具有显著差异性;其中,牛、绵羊和山羊PRNP基因多态性与BSE、瘙痒病显著相关;牛、绵羊和山羊PRNP基因多态性与生产性能有密切相关性。为此,本文从反刍家畜PRNP基因结构比较、反刍家畜PRNP基因多态性研究、反刍家畜PRNP多态性与疾病的关系、反刍家畜PRNP多态性与生产性能的关系等四方面进行综述,以期为反刍家畜优良个体及品种的高效选育提供理论参考。
反刍家畜;朊蛋白基因(PRNP);多态性;遗传效应;性状
作为人兽共患疾病的典型代表,传染性海绵样脑病(transmissible spongiform encephalopathies,TSEs)因其高传染性与致死性对畜牧产业乃至整个社会造成了极大的危害[1]。作为调控哺乳动物TSEs的主要基因,朊蛋白基因(prion protein gene,PRNP)突变或其编码蛋白构象改变,会引起朊病毒疾病的发生。目前,已有研究证实反刍家畜PRNP基因存在丰富的遗传多态性,同时这些遗传变异位点在这些物种中又具有显著差异性;其中牛[2]、山羊[3]和绵羊[4]的PRNP基因多个多态性位点与TSEs的易感性及潜伏期有显著的相关关系。典型的朊病毒病包括人的克雅氏病、吉斯特曼斯特劳斯综合征、致死性家族性失眠症、散发型致死性失眠病、库鲁病、变异型克雅氏病、牛传染性海绵状脑病或称疯牛病和羊瘙痒病等[5]。疾病的发生会影响家畜的生长速度,产子数以及其他生产性能等,从而造成了严重的经济生产损失[6]。随着研究的逐渐深入,也有研究发现PRNP基因多态性可以影响未感染TSEs的健康动物的表型性状[7,8],因此,研究PRNP基因多态性对家畜生产性能的提高具有重要意义。
为此,本文对反刍家畜PRNP基因结构比较、反刍家畜PRNP基因多态性研究、反刍家畜PRNP多态性与疾病的关系、反刍家畜PRNP多态性与生产性能的关系等方面进行综述,以期为分子标记辅助优良个体选择及家畜育种研究提供了重要思路与理论支持。
1 反刍家畜PRNP基因结构的比较
由于PRNP基因广泛存在于动物体内,目前研究人员已对多种动物的PRNP基因进行了研究。在常见反刍家畜中,例如山羊、绵羊、家牛、瘤牛、水牛、牦牛等,PRNP基因大多被定位于家畜13号染色体(牦牛未定位),大多含有三个或五个外显子,PRNP 基因的开放阅读框(ORF)都编码一个约由250个氨基酸组成的PrP蛋白,基因结构较简单(表1)。此外,在已研究的哺乳动物中,PRNP基因ORF中的DNA序列和PrP氨基酸序列的相似性分别达到90%和95%以上[9]。由此可见,PRNP基因具有高度保守性,这也为朊蛋白跨物种传播提供了分子基础。
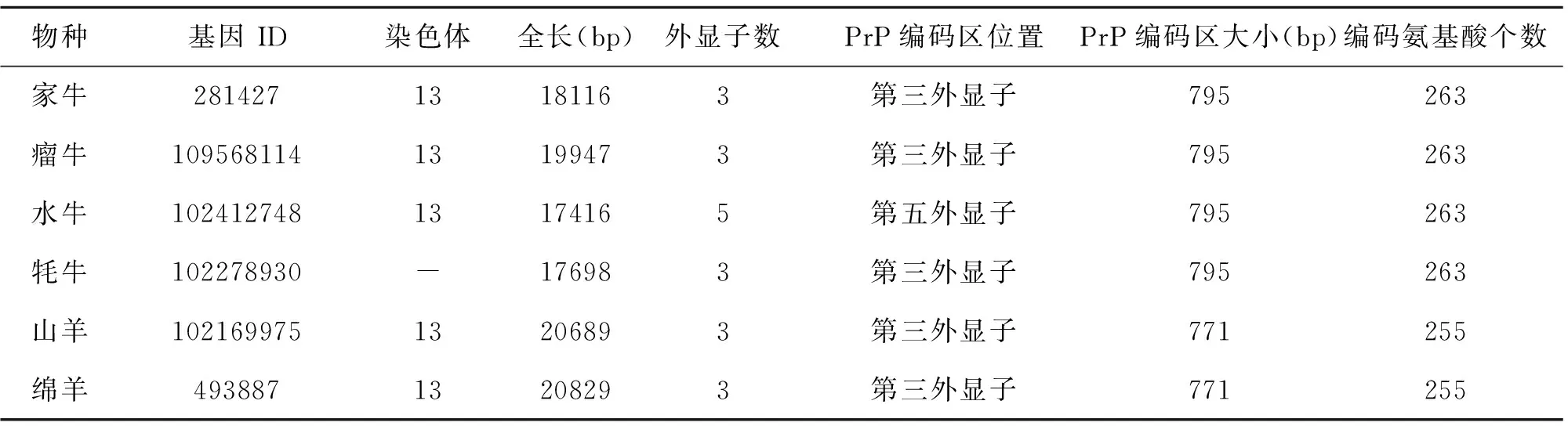
表1 常见反刍家畜PRNP基因结构统计表
2 反刍家畜PRNP基因多态性研究进展
虽然PRNP基因在反刍家畜之间具有高度同源性,但该基因在不同物种之间多态性却具有很大差异性,这也是PRNP基因多态性一直是研究热点的原因之一。目前,反刍家畜PRNP基因已被报道的多态性位点包括单核苷酸多态性(SNPs)和插入/缺失(indel)突变,拷贝数变异(CNV)却未见报道。
2.1 反刍家畜PRNP基因SNP多态性研究进展
反刍家畜PRNP基因中存在大量的SNP位点,且不同品种间的SNP多态性差异较大。早在2010年,Shimogiri等通过对mythun,日本黑牛以及蒙古牛群体进行检测,发现K3T和S154N两个无义突变[10]。Xi等通过对125头大额牛的PRNP基因编码区序列进行检测,共发现10个SNP位点,包括6个同义突变(C60T,G75A,A108T,G126A,C357T和C678T)和4个非同义突变(C8A,G145A,G461A和C756G),相对应的氨基酸上的改变(T3K,G49S9,N154S和I252M)也得到了验证[11]。与此同时,Choi等首次对300头Hanwoo牛(韩国),泽西牛,日本黑牛等群体的PRNP基因编码区序列进行检测,发现4个同义突变(G234A,C555T,C576T,和C630T) 和2个非同义突变[12]。在中国延边牛和中国草原红牛群体中,PRNP基因也存在3个非同义突变造成了氨基酸的改变(K119N,S154N和 M177V)和一个沉默突变(A234G)[13]。
在安托利亚水牛及摩拉水牛PRNP基因编码区检测到3个同义突变(126,234及285位)与一个非同义突变(322位,G108S)。其中,126位存在G/A/T三种碱基[14];水牛PRNP基因也具有较高多态性,但牦牛PRNP基因多态性的相关研究还未见报道。
绵羊PRNP基因中有大量SNP位点,例如,密码子85 (G/R),112 (M/T/I),116 (A/P),127(G/A/V/S),136 (A/V/T),137 (M/T),138 (S/N),141(L/F),143 (H/R),151 (R/C),152 (Y/F),154 (R/H),167 (R/S),168 (P/L),171 (Q/R/H/K),175 (Q/E),176(K/N),180 (H/Y),189 (G/L/R),195 (T/S),196 (T/S),211 (R/Q)和161 (P/S)[15-19]。Lan等对来自中国16个地方品种的486只绵羊进行了检测,共检测到154位密码子(R或H)和171 位密码子(具有4种突变,分别编码Q,R,H或K)具有多态性。在所有检测品种中,136位密码子均为纯合AA,未检测到A/V突变。此外,在所有检测品种中,密码子21,101,112,127,138,141,143,146,153和189 均具有多态性。密码子171位的优势等位基因为Q(频率高达88.68%)[20]。
在山羊群体中,有大量SNP位点,分别位于密码子:V21A,G22C,L23P,G37V,G42A,G49S,P63L,W102G,T110N,T110P,G127S,L133Q,M137I,I142M,I142T,H143R,N146S,N146D,R151H,R154H,P168Q,T194P,F201L,R211Q,R211G,I218L,T219I,Q220H,Q222K和P240S[21-31]。Zhou等对来自中国11个地方品种的337只山羊羊进行了检测,共在山羊PRNP基因上检测到10种氨基酸的多态性(分别位于密码子102,127,143,146,154,211,218,219,222和240)[32]。
2.2 反刍家畜PRNP基因indel多态性研究进展
反刍家畜PRNP基因中除了存在大量的SNPs位点,还存在一些indel位点,如表2所示。
目前,关于人类PRNP基因CNV研究有极少报道,例如:Ana等[33]通过对1147名克雅氏病人进行PRNP CNV扫描,经多重检验后发现CNVs >100,>500,或者 >1000 kb,与人克雅氏疾病的发生并无显著关联性。但关于反刍家畜PRNP基因拷贝数变异(CNV)却未见报道,反刍动物PRNP 是否存在CNV变异及其遗传效应仍有待挖掘。
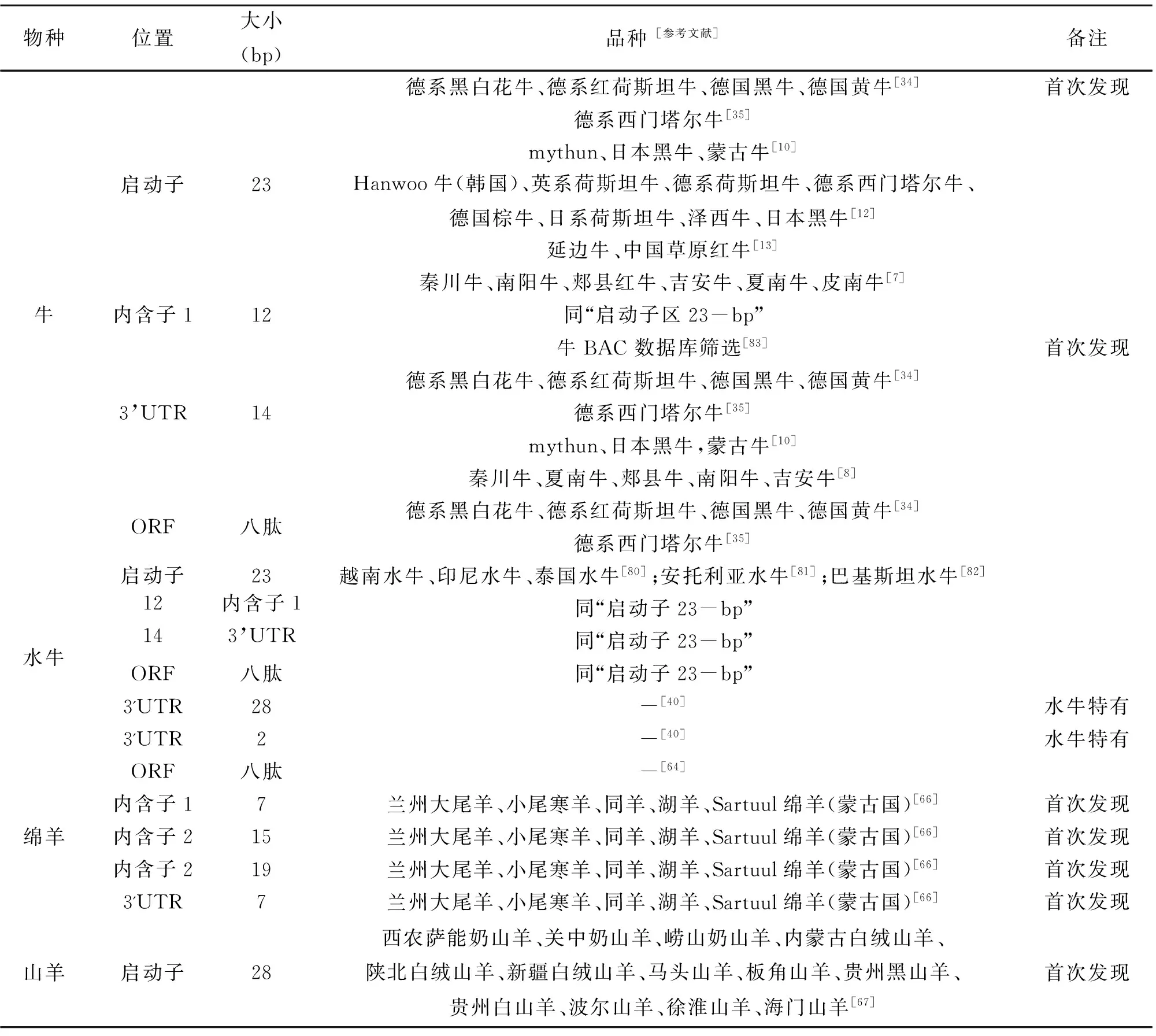
表2 常见反刍家畜PRNP基因indel位点统计表
3 反刍家畜PRNP多态性与疾病的关系
朊蛋白基因(prion protein gene,PRNP)突变或其编码蛋白构象发生改变,是引发哺乳动物TSEs的主要原因。Sander等[34,35]分析了43头已感染BSE的德国肉牛与48头健康的德国肉牛,也是第一次比较了23-bp indel和12-bp indel这两个indel多态性位点与BSE抗病性的相关关系,结果发现PRNP基因多态性与BSE有显著相关,并且也可以调节BSE的潜伏期,与疯牛病的易感性相关联。并且,他们发现这两个indel位点可以影响转录因子RP58和Sp1的结合位点。Xue等通过对日本黑牛的研究,发现转录因子Sp1和RP58可能对PRNP基因的表达有负反馈调节作用[36]。同时,Kashkevich等(2007)研究PRNP基因启动区域的多态性,发现通过降低PRNP基因的表达可以增强疯牛病的抗性[37]。而Neibergs等[38]报道了开放阅读框并不影响疯牛病的易感性。Hreko等研究发现,PRNP基因启动子区的23-bp缺失的纯合子插入基因型似乎已经对疯牛病起保护作用[39]。此外,由于水牛与家牛PrP的表达量不同,因此牛与水牛对疯牛病的易感性也明显不同[40-41]。至今还未在水牛群体中见到爆发疯牛病。大量研究表明牛PRNP启动子区23-bp及第一内含子12-bp的多态性与疯牛病感染性及抗性密切相关,但在安纳托利亚水牛及摩拉水牛中只发现了插入/插入(II)型,未检测到突变型,推测II型为抗性基因型[42]。
绵羊PRNP基因多态性研究多集中于外显子区域,尤其是第三外显子且突变多以单核苷酸突变为主。136[43],141[44],146[45],154[46]和171[47]位密码子的变异被证实与绵羊瘙痒病的发生或病症密切相关。研究表明,在患有TSE的病羊中,其136 位点(A-V)154 位点(R-H)和171 位点(Q-R/H)均会发生突变。其中,Q171R多态性对羊瘙痒病的感染具有很强的抗性而A136V多态性则极易感染。并且,基因型ARR/ARR 或ARR/ARQ 是抗性基因型,而ARQ/ARQ、VRQ/ARQ、AHQ/VRH 等其它基因型羊则为敏感型[48-51]。同时,一些非编码区域的突变也被报道影响绵羊瘙痒病的易感性[52]。
类似地,山羊PRNP基因的多态性,如I142M,H143R,N146S/D,R154H,R211Q和Q222K也对羊瘙痒病的感染表现出明显的抗性[5]。自2001年开始,欧盟各国开始培育有抗性基因的羊,其中ARR/ARR纯合子具有最高的抗羊瘙痒病的能力,并且这些纯合子绵羊的产量、生长、繁殖和健康都和其他羊没有明显的差异[53]。随着家畜抗TSEs分子育种工作在欧盟各国成功开展,这些著名的致病性位点经过人工选择逐渐被淘汰,绵羊瘙痒病和疯牛病等神经退行性疾病的爆发也基本已得到控制。
4 PRNP多态性与生产性能的关系
生产性能是决定畜牧养殖业效益的主要指标,包括家畜生长性状及繁殖性能。除了参与调控TSEs疾病,PRNP基因多态性也被证实与动物的一些表型性状显著相关。例如,绵羊PRNP基因的VRQ和ARQ等位基因变异体(在密码子136、154和171的多态性位点)与绵羊瘙痒病的高易感性呈显著相关,导致绵羊的生产性能显著降低从而造成了严重的经济生产损失[6]。与此同时,也有研究表明PRNP基因多态性可以影响未感染TSEs的健康动物的表型性状。
在健康牛群体中,PRNP八肽重复序列多态性被报道与荷斯坦奶牛的产奶性状密切相关[54]。在我国地方黄牛群体中,PRNP基因多样性与生产性能的关系也被揭示。Yang[7-8]等首次在中国良种黄牛(秦川牛,南阳牛,郏县红牛,吉安牛,夏南牛和皮南牛)群体中,验证了PRNP基因3′ UTR区14-bp indel,启动子区23-bp indel和第一内含子12-bp indel突变与牛的生长性状显著相关。例如,3′ UTR区14-bp indel与秦川牛的体长,夏南牛的体重和腰围显著相关(P<0.05)。南阳牛在23-bp indel位点上基因型为缺失/缺失(DD)基因型的个体以及12-bp indel位点基因型为插入/插入(II)基因型的个体表现出了更优良的表型性状,在六个研究的牛品种中南阳牛具有较低的杂合子(ID)单倍型频率[8]。此外,PRNP启动子区23-bp indel显著影响南阳牛的体长和胸围,第一内含子12-bp indel突变也与吉安牛的管围密切相关[7]。
在健康绵羊群体中,PRNP基因多态性被报道影响羔羊生长性状[55-57],母绵羊的繁殖力[58-60]和产奶量[61-62]等生产性能。Sawalha等研究发现,携带ARQ等位基因的苏格兰黑面母绵羊往往具有较强的季节性脂肪组织动员能力[63]。同时,他们也发现171位密码子的多态性与萨福克绵羊的产羔性状显著相关[25]。Allais-Bonnet[64]等人也发现PRNP基因型的差异(例如,ARQ/ARQ和 ARR/ARQ)及八肽重复序列多态性可以导致Latxa绵羊产奶性能(产奶量,奶蛋白,奶脂含量)的差异。还有研究表明PRNP基因型与Texel羊的产仔数和135日龄的体重有显著相关[65]。先前我们课题组在来自中国和蒙古5个品种的768名健康绵羊个体PRNP基因中共发现了四种新的indel多态性:第一内含子7-bp(I1-7bp),第二内含子15-bp(I2-15 bp),第二内含子19-bp(I2-19 bp)和3 'UTR-7 bp(3'UTR-7 bp)。并且,这4种indel与13个不同的生长性状(例如,毛长,背高,胸围等)呈显著相关(P<0.05)[66]。尤其是I2-15 bp对小尾寒羊(母羊)的胸宽(P= 0.001,DD为优势基因型),3'UTR-7 bp对湖羊的胸围(P= 0.003,DD为优势基因型),12-19 bp 对小尾寒羊(公羊)的胸围指数(P=1.122E-4,DD为优势基因型)以及同羊的尾长(P= 0.001,II为优势基因型),呈现极显著关联性(P<0.01)[66]。
在健康山羊群体中,山羊PRNP基因启动子区28-bp的indel突变与内蒙古白绒山羊的平均体重,1岁羊平均体重以及3岁羊毛厚度显著相关,并且II型均为这三种性状的优势基因型[67]。此外,该 indel 突变也与西农萨能奶山羊的平均产奶量密切相关,II型个体产奶量显著高于杂合型(ID)个体,但ID型个体夜间总固体量要显著高于II型个体[67]。此外,Lan等通过对来自4个绵羊品种(西农萨能奶山羊,内蒙古白绒山羊,新疆绒山羊,陕北白绒山羊)2002只绵羊的PRNP基因进行多态性位点扫描,发现第42位密码子的SNP位点(42CCG>42CCA)与7岁内蒙古白绒山羊的体重显著相关,并且GG型个体体重要显著优于AA基因型个体[68]。与此同时,该位点还与内蒙古白绒山羊的产绒量和毛长显著相关。此外,在西农萨能奶山羊中,GG型个体的早、晚奶密度以及奶中非脂质固体含量要显著优于其他基因型。总体而言,GG型萨能奶山羊所产奶质量往往要优于其他基因型[68]。
5 展望
对于PRNP基因多态性与TSEs发病机理的关系,Telling等(1996年)学者认为PRNP基因点突变的发生增加了不稳定的中间分子的数量,同时加快了PrPc向PrPsc的转换速率,从而形成稳定性更好的PrPsc构象[69]。此外,有研究证明Pr P在形成二级结构过程中,该区域很容易发生α螺旋转变为β 折叠,增加了发病机率[70]解释了PRNP 多态性与TSEs发病机理的关系。
在医学领域中已有报道称PrPc参与胚胎自我修复,组织分化和癌干细胞的生成等过程[71-72],这为朊蛋白基因调控动物的生产性状提供了理论基础。先前研究报道DNA序列上碱基的突变可能会改变mRNA的稳定性,以及mRNA加工和成熟过程,进一步影响等位基因的表达和翻译后肽链的折叠[73]。因此,我们预测PRNP这些indel突变序列中可能存在其他生长主效基因转录因子的结合位点,使得这些生长基因的表达受到影响[74]。另外,有报道称PRNP基因3′ UTR区的indel多态性可以通过微小RNA介导的转录后机制来调控性状或疾病易感性[75]。此外,基因间的相互作用也可能是PRNP基因调控生长性状的原因之一。已有研究证明在绵羊群体中,PRNP多态性位点往往与其临近同源基因-叠朊蛋白基因(PRND)的多态性位点相互关联[76],而我们先前的研究结果表明,绵羊PRND基因编码区上游的20-bp indel突变与湖羊的管围指数显著相关[77]。此外,绵羊PRNP和GnRH基因也被报道可能存在连锁关系,影响绵羊性成熟时的体重,对乳腺分化、母性行为、免疫特征及细胞凋亡都存在影响[78]。然而,这其中确切的分子调控机制还需要继续探究。
迄今为止,我国还未见有关疯牛病和瘙痒病大规模爆发的报道,且相关研究表明我国部分地方品种感染TSEs的可能性较低[79],因此如何在健康群体中高效准确的筛选优良个体及培育优良畜种成为养殖业的关键。因DNA标记具有标记范围广,标记数量不受限制,遗传相对稳定等优点,分子标记辅助选择(MAS)在畜牧业被广泛的应用,发展也十分迅速。目前已有应用的DNA分子标记包括:RFLP标记、SSR标记、indel 标记以及SNP标记等多种标记方法,这些方法能够加快育种改良进程、减少生产成本。与此同时,上述大量研究已表明PRNP基因在反刍家畜中具有较高的多态性,可作为MAS选择靶向基因。伴随着畜牧产业的快速发展,相信未来会有更多的研究关注PRNP的遗传多样性与动物经济性状的关系,从而为分子标记辅助选择(MAS)在畜牧业中的广泛应用提供新思路与理论依据,加快家畜优良经济品种的选育进程。
[1] 席冬梅,刘 情,于虹漫,等. 牛羊朊蛋白基因( PRNP) 多态性与抗病性的研究进展[J]. 云南农业大学学报, 2011, 26(3): 418-425.
[2] Goldmann W, Martin T, Foster J, et al. Novel polymorphisms in the caprine prp gene. A codon 142 mutation associated with scrapie incubation period [J]. Gen Virol, 1996, 77 (11): 2885-2891.
[3] Goldmann W. PrP genetics in ruminant transmissible spongiform encephalopathies[J]. Vet Res. 2008, 39(4): 30.
[4] Goldmann W, Hunter N, Smith G, et al. PrP genotype and agent effects in scrapie: change in allelicinteraction with different isolates of agents in sheep, a natural host of scrapie [J]. Gen Virol. 1994, 75 (5): 989-995.
[5] 张会侠,师 润,李朝阳. 朊病毒疾病将如何发展[J]. 科学通报, 2017, 62: 16-24.
[6] Ioannides IM, Mavrogenis AP, Papachristoforou C. Analysis of PrP genotypes in relation to reproductive and production traits in Chios sheep [J]. Livest Sci, 2009, 122(2-3): 296-301.
[7] Yang Q, Zhang SH, Liu LL, et al. The evaluation of 23-bp and 12-bp insertion /deletion within the PRNP gene and their effects on growth traits in healthy Chinese native cattle breeds[J]. Journal of Applied Animal Research, 2017, dio: 10.1080/09712119.2017.1348950.
[8] Yang Q, Zhang SH, Liu LL, et al. Application of mathematical expectation (ME) strategy for detecting low frequency mutations: an example for evaluating 14 bp insertion/deletion (indel) within the bovine PRNP gene[J]. Prion, 2016, 10: 409-419.
[9] Lee IY, Westaway D, Smit AF, et al. Complete genomic sequence and analysis of the prion protein gene region from three mammalian species. Genome Research, 1998, 8 (10): 1022-1037.
[10] Shimogiri T, Msalya G, Myint SL, et al. Allele distributions and frequencies of the six prion protein gene (PRNP) polymorphisms in Asian native cattle, Japanese breeds, and mythun (Bos frontalis)[J]. Biochem Genet, 2010, 48(9-10):829-839.
[11] Xi D, Liu Q, Guo J, et al. Genetic variability of the coding region for the prion protein gene (PRNP) in gayal (Bos frontalis) [J]. Mol Biol Rep, 2012, 39(2):2011-2020.
[12] Choi S, Woo HJ, Lee J. Sequence variations of the bovine prion protein gene (PRNP) in native Korean Hanwoo cattle[J]. J Vet Sci, 2012, 13(2):127-137.
[13] Qin LH, Zhao YM, Bao YH, et al. Polymorphism of the prion protein gene (PRNP) in two Chinese indigenous cattle breeds[J]. Mol Biol Rep, 2011, 38(6):4197-4204.
[14] Yaman Y, ün C. Nucleotide and octapeptide-repeat variations of the prion protein coding gene (PRNP) in Anatolian, Murrah, and crossbred water buffaloes[J]. Trop Anim Health Prod, 2017, doi: 10.1007/s11250-017-1471-9.
[15] Heaton MP, Leymaster KA, Freking BA, et al. Prion gene sequence variation within diverse groups of U.S. sheep, beef cattle, and deer[J]. Mamm Genome: Off J Int Mamm Genome Soc, 2003, 14(11):765-777.
[16] Belt PB, Muileman IH, Schreuder BE, et al. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie[J]. J Gen Virol, 1995, 76(3):509-517.
[17] Billinis C, Psychas V, Leontides L, et al. Prion protein gene polymorphisms in healthy and scrapie-affected sheep in Greece[J]. J Gen Virol, 2004, 85(2):547-554.
[18] Zhang L, Li N, Fan B, et al. PRNP polymorphisms in Chinese ovine, caprine and bovine breeds[J]. Anim Genet, 2004, 35(6):457-461.
[19] Lan Z, Wang ZL, Liu Y, et al. Prion protein ene (PRNP) polymorphisms in Xinjiang local sheep breeds in China[J]. Arch Virol, 2006, 151(10):2095-2101.
[20] Lan Z, Li J, Sun C, et al. Allelic variants of PRNP in 16 Chinese local sheep breeds[J]. Arch Virol, 2014, 159(8):2141-2144.
[21] Goldmann W, Martin T, Foster J, et al. Novel polymorphisms in the caprine PrP gene: a codon 142 mutation associated with scrapie incubation period[J]. J Gen Virol, 1996, 77(11):2885-2891.
[22] Goldmann W, Chong A, Foster J, et al. The shortest known prion protein gene allele occurs in goats, has only three octapeptide repeats and is non-pathogenic[J]. J Gen Virol, 1998, 79(12): 3173-3176.
[23] Goldmann W, Perucchini M, Smith A, et al. Genetic variability of the PrP gene in a goat herd in the UK[J]. Vet Rec, 2004, 155(6):177-178.
[24] Goldmann W, Ryan K, Stewart P, et al. Caprine prion gene polymorphisms are associated with decreased incidence of classical scrapie in goat herds in the United Kingdom[J]. Vet Res, 2011, 42(1):110.
[25] Billinis C, Panagiotidis CH, Psychas V, et al. Prion protein gene polymorphisms in natural goat scrapie[J]. J Gen Virol,2002, 83(3): 713-721.
[26] Kurosaki Y, Ishiguro N, Horiuchi M, et al. Polymorphisms of caprine PrP gene detected in Japan[J]. J Vet Med Sci, 2005, 67(3):321-323.
[27] Acutis PL, Bossers A, Priem J, et al. Identification of prion protein gene polymorphisms in goats from Italian scrapie outbreaks[J]. J Gen Virol,2006, 87(4):1029-1033.
[28] Acutis PL, Colussi S, Santagada G, et al. Genetic variability of the PRNP gene in goat breeds from Northern and Southern Italy[J]. J Appl Microbiol. 2008; 104(6):1782-1789.
[29] Babar ME, Abdullah M, Nadeem A, et al. Prion protein gene polymorphisms in four goat breeds of Pakistan[J]. Mol Biol Rep, 2009, 36(1):141-144.
[30] Hussain A, Babar ME, Imran M, et al. Detection of four novel polymorphisms in PrP gene of Pakistani sheep (Damani and Hashtnagri) and goats (Kamori and Local Hairy) breeds[J]. Virol J, 2011, 8:246.
[31] Papasavva-Stylianou P, Windl O, Saunders G, et al. PrP gene polymorphisms in Cyprus goats and their association with resistance or susceptibility to natural scrapie[J]. Vet J, 2011, 187(2):245-250.
[32] Zhou R, Li X, Xi J, et al. Genetic variability of PRNP in Chinese indigenous goats[J]. Biochem Genet, 2013, 51(3-4):211-22.
[33] Lukic A, Uphill J, Brown CA, et al. Rare structural genetic variation in human prion diseases[J]. Neurobiol Aging, 2015, 36(5):2004.e1-8.
[34] Sander P, Hamann H, Pfeiffer I, et al. Analysis of sequence variability of bovine prion protein gene (PRNP) in German cattle breeds[J]. Neurogenetics, 2004, 5(1): 19-25.
[35] Sander P, Hamann H, Drgemüller C, et al. Bovine prion protein (PRNP): promoter polymorphisms modulated PRNP expression and may be responsible for differences in bovine spongiform encephalopathy susceptibility[J]. Biol Chem, 2005, 280(45): 37408-37414.
[36] Xue G, Sakudo A, Kim CK, et al. Coordinate regulation of bovine prion protein gene promoter activity by two Sp1 binding site polymorphisms[J]. Biochem Biophys Res Commun, 2008, 372(4):530-535.
[37] Kashkevich K, Humeny A, Ziegler U, et al. Fuctional relevance of DNA polymorphisms within the promoter region of the prion protein gene and their association to BSE infection[J]. FASEB J, 2007, 21(7):1547-1555.
[38] Neibergs HL, Ryan AM, Womack JE, et al. Polymorphism analysis of the prion gene in BSE-affected and unaffected cattle[J]. Anim Genet, 1994, 25(5): 313-317.
[39] Hresko S, Mojzis M, Tkacikova L. Prion protein gene polymorphism in healthy and BSE-affected Slovak cattle[J]. Journal of Applied Genetics, 2009, 50(4): 371-374.
[40] Zhao H, Wang S, Guo L, et al. Fixed differences in the 3'UTR of buffalo PRNP gene provide binding sites for miRNAs post-transcriptional regulation[J]. Oncotarget, 2017, 8(28):46006-46019.
[41] Zhao H, Du Y, Chen S, et al. The prion protein gene polymorphisms associated with bovine spongiform encephalopathy susceptibility differ significantly between cattle and buffalo[J]. Infect Genet Evol, 2015, 36:531-538.
[42] Yaman Y, Karada O, ün C. Investigation of the prion protein gene (PRNP) polymorphisms in Anatolian, Murrah, and crossbred water buffaloes (Bubalus bubalis)[J]. Trop Anim Health Prod, 2017, 49(2):427-430.
[43] Caroline P. de Andrade, Laura L, et al. Development of a real-time polymerase chain reaction assay for single nucleotide polymorphism genotyping codons 136, 154, and 171 of the prnp gene and application to Brazilian sheep herds[J]. Journal of Veterinary Diagnostic Investigation, 2013, 25(1): 120-124.
[44] Konold T, Phelan LJ, Donnachie BR, et al. Codon 141 polymorphisms of the ovine prion protein gene affect the phenotype of classical scrapie transmitted from goats to sheep[J]. BMC Veterinary research, 2017, 13(1): 122.
[45] Papasavva-Stylianou P, Simmons MM, Ortiz-Pelaez A, et al. The effect of polymorphisms at codon 146 of the goat PRNP gene on susceptibility to challenge with classical scrapie by different routes[J]. Journal of Virology, 2017, doi: 10.1128/JVI.01142-17.
[46] Seabury CM, Derr JN. Identification of a novel ovine PrP polymorphism and scrapie-resistant genotypes for St. Croix White and a related composite breed[J]. Cytogenetic and Genome Research,2003, 102: 85-88.
[47] Zabavnik J, Cotman M, Juntes P, et al. A decade of using small-to-medium throughput allele discrimination assay to determine prion protein gene (Prnp) genotypes in sheep in Slovenia[J]. Journal of Veterinary Diagnostic Investigation, 2017, doi: 10.1177/1040638717723946.
[48] Buschmann A,Lühken G,Schultz J,et al. Neuronal accumulation of abnormal prion protein in sheep carrying a scrapie-resistant genotype(PrPARR/ARR)[J]. J Gen Virol, 2004, 85(9): 2727-2733.
[49] Hamir A N, Kunkle R A, Richt J A, et al. Experimental transmission of sheep scrapie by intracerebral and oral routes to genetically susceptible Suffolk sheep in the United States[J]. J Vet Diagn Invest, 2005, 17(1): 3-9.
[50] Baylis M,McIntyre K M. Transmissible spongiform encephalopathies: scrapie control under new strain[J]. Nature, 2004, 432(7019): 810-811.
[51] 肖妍,董志珍,栾慎顺,等. 我国部分地区传染性羊痒病基因型的分布情况调查[J]. 中国动物检疫, 2011, 28(12); 45-47.
[52] Saunders GC, Cawthraw S, Mountjoy SJ, et al. Ovine PRNP untranslated region and promoter haplotype diversity[J]. Journal of General Virology, 2009, 90: 1289-1293.
[53] Mesquita P, Batista M, Marques MR, et al. Prion-liked Doppel gene polymorphisms and scrapie susceptibility in portuguese sheep breeds[J]. Animal Genetics, 2010, 41(3): 311-314.
[54] Walawski K, Czarnik U. Prion octapeptide-repeat polymorphism in Polish Black-and-White cattle [J]. Appl Genet, 2003, 44(2): 191-195.
[55] Isler BJ, Freking BA, Thallman RM, et al. Evaluation of associations between prion haplotypes and growth, carcass, and meat quality traits in a Dorset x Romanov sheep population[J]. Journal of Animal Science, 2006, 84: 783-788.
[56] Sawalha RM, Brotherstone S, Man WYN, et al. Associations of polymorphisms of the ovine prion protein gene with growth, carcass, and computerized tomography traits in Scottish Blackface lambs[J]. Journal of Animal Science, 2007, 85: 632-640.
[57] Sawalha RM, Villanueva B, Brotherstone S, et al. Prediction of prion protein genotype and association of this genotype with lamb performance traits of Suffolk sheep[J]. American Society of Animal Science, 2010, 88(2): 428-434.
[58] Ponz R, Tejedor MT, Monteagudo LV, et al. Scrapie resistance alleles are not associated with lower prolificity in Rasa Aragonesa sheep[J]. Research in Veterinary Science, 2006, 81: 37-39.
[59] Casellas J, Caja G, Bach R, et al. 2007. Association analyses between the prion protein locus and reproductive and lamb weight traits in Ripollesa sheep[J]. Journal of Animal Science, 2007, 85: 592-597.
[60] Guan F, Pan L, Li J, et al. Polymorphisms of the prion protein gene and their effects on litter size and risk evaluation for scrapie in Chinese Hu sheep[J]. Virus Genes, 2011, 43(1): 147-152.
[61] álvarez L, Gutiérrez-Gil B, San Primitivo F, et al. Influence of prion protein genotypes on milk production traits in Spanish Churra sheep[J]. Journal of Dairy Science, 2006, 89: 1784-1791.
[62] Psifidi A, Basdagianni Z, Dovas CI, et al. Characterization of the PRNP gene locus in Chios dairy sheep and its association with milk production and reproduction traits[J]. Animal Genetics, 2011,42(4): 406-414.
[63] Sawalha RM, Brotherstone S, Lambe NR, et al. Association of the prion protein gene with individual tissue weights in Scottish Blackface sheep[J]. Journal of Animal Science, 2008, 86: 1737-1746.
[64] Allais-Bonnet A, Castille J, Pannetier M, et al. A specific role for PRND in goat foetal Leydig cells is suggested by prion family gene expression during gonad development in goats and mice[J]. FEBS Open Bio, 2016, 6(1): 4-15.
[65] Brandsma JH, Janss LLG, Visscher AH. Association between PrP genotypes and littersize and 135 days weight in Texel sheep [J]. Liv. Produc Sci, 2004, 85: 59-64.
[66] Jie Li, Sarantsetseg Erdenee, Shaoli Zhang, et al. Genetic effects of PRNP gene insertion/deletion (indel) on phenotypic traits in sheep[J]. Prion, 2017, doi: 10.1080/19336896.2017.1405886.
[67] Lan XY, Zhao HY, Li ZJ, et al. A novel 28-bp insertion-deletion polymorphism within goat PRNP gene and its association with production traits in Chinese native breeds [J]. Genome, 2012, 55(7): 547-552.
[68] Lan X, Zhao H, Wu C, et al.Analysis of genetic variability at codon 42 within caprine prion protein gene in relation to production traits in Chinese domestic breeds[J]. Mol Biol Rep, 2012 ,39(4):4981-4988.
[69] Telling G C, Parchi P, DeArmond S J, et al. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity[J]. Science, 1996, 274(5295):2079-2082.
[70] 杨建民,赵德明,郝永新,等. 奶牛朊病毒基因克隆与序列分析[J]. 畜牧兽医学报, 2004, 35(6): 685-688.
[71] Rousset M, Leturque A, Thenet S. The nucleo-junctional interplay of the cellular prion protein: A new partner in cancer-related signaling pathways? [J] Prion, 2016, 10(2): 143-152.
[72] Santos TG, Lopes MH, Martins VR. Targeting prion protein interactions in cancer[J]. Prion, 2015, 9(3): 165-173.
[73] Komar AA. Genetics. SNPs, silent but not invisible[J]. Science. 2007, 315: 466-467.
[74] Hou JX, An XP, Song YX, et al. Two mutations in the caprine MTHFR 3’ UTR regulated by microRNAs are associated with milk production traits[J]. PLoS One, 2015, 10(7): e0133015.
[75] Peletto S, Bertolini S, Maniaci MG, et al. Association of an indel polymorphism in the 3’UTR of the caprine SPRN gene with the scrapie positivity in the central nervous system[J]. Journal of General Virology, 2012, 93: 1620-1623.
[76] Mesquita P, Garcia V, Marques MR, et al. The prion-related protein (testis-specific) gene (PRNT) is highly polymorphic in Portuguese sheep[J]. Animal Genetics, 2015, 47: 128-132.
[77] Li J, Zhu XC, Ma L, et al. Detection of a new 20bp insertion/deletion (indel) within sheep PRND gene using mathematical expectation (ME) method. Prion. 2017,11(2): 143-150.Sweeney T, Hanrahan JP. The evidence of associations between prion protein genotype and production, reproduction, and health traits in sheep[J]. Vet Res, 2008, 39(4): 28?46.
[78] Sweeney T, Hanrahan JP. The evidence of associations between prion protein genotype and production, reproduction, and health traits in sheep[J]. Vet Res, 2008, 39(4): 28-46.
[79] 朱庆丰,刁俊超,潘 磊,等. 湖羊PRNP 多态性及其对产羔数影响和抗痒病风险评价[J]. 中国预防兽医学报,2012, 34(10):802-806.
[80] Uchida L, Heriyanto A, Thongchai C, et al. Genetic diversity in the prion protein gene (PRNP) of domestic cattle and water buffaloes in Vietnam, Indonesia and Thailand[J]. J Vet Med Sci, 2014 , 76(7): 1001-1008.
[81] Oztabak K, Ozkan E, Soysal I, et al. Detection of prion gene promoter and intron1 indel polymorphisms in Anatolian water buffalo (Bubalus bubalis)[J]. J Anim Breed Genet, 2009, 126(6): 463-467.
[82] Imran M, Mahmood S, Babar ME, et al. PRNP gene variation in Pakistani cattle and buffaloes[J]. Gene, 2012, 505(1): 180-185.
[83] Hills D, Comincin S, Schlaepfer J, et al. Complete genomic sequence of the bovine prion gene (PRNP) and its polymorphism in its promoter region [J]. Animal Genetics, 2001, 32(4): 231-232.
