Screening of cellulose decomposing fungi in sandy dune soil of Horqin Sandy Land
2015-12-19ShaoKunWangXueYongZhaoXiaoAnZuoXinPingLiuHaoQuWeiMaoJianYingYun
ShaoKun Wang ,XueYong Zhao ,XiaoAn Zuo ,XinPing Liu ,Hao Qu ,2,Wei Mao ,2,JianYing Yun
1.Naiman Desertification Research Station,Cold and Arid Regions Environmental and Engineering Research Institute,Chinese Academy of Sciences,Lanzhou,Gansu 730000,China
2.University of Chinese Academy of Sciences,Beijing 100039,China
1 Introduction
Cellulose is the most renewable biological energy source in the world.It is estimated that around 150–200 billion tons of dry plant biomass is produced by photosynthesis every year,of which 50% is cellulose(Kubiceket al.,1993;Lyndet al.,2002).Environmental stress and energy shortages can be relieved by proper treatment and effective utilization of cellulose,which can produce substances such as sugar,alcohol,and organic fertilizer during its decomposition(Regnellet al.,2014).Cellulose decomposition is the key point of the natural carbon cycle,and it plays an important role in nutrient cycling and maintaining ecological functions in ecosystems(Schwarz,2001;Chapin IIIet al.,2011).Cellulose decomposers are a type of microbes that can decompose cellulose by biological hydrolyzation;they include bacteria,actinomycetes,and fungi.Most of the cellulose decomposers isolated from the natural environment are fungi,such asTrichoderma,Aspergillus,Penicillium,andFusarium(Bisaria and Ghose,1981;Panagiotouet al.,2003;Wenet al.,2005).
Researches on isolation and screening of cellulose decomposers are increasing with the development of screening techniques and the demand for cellulose metabolites.Investigators have isolated and screened strains or/and microbial communities which can produce highly efficient cellulolytic enzymes in forests(Štursováet al.,2012),farmland(Houet al.,2013),grasslands(Luet al.,2011),compost(Niuet al.,2005),and animal stomachs(Fanet al.,2012).Cellulose decomposers are an important functional microbial group in sandy land ecosystems,because of their ability to decompose litter(Kamolmanitet al.,2013).Litter can maintain diversity and stability in sandy land ecosystems by adjusting soil climate,improving soil nutrients,and increasing vegetation biomass(Quet al.,2011).Cellulose decomposers were detected in sandy dune soils in previous research(Wanget al.,2013),which enabled screening of efficient cellulose decomposing fungi in the Horqin Sandy Land.We demonstrated that isolation and screening of highly efficient cellulose decomposing fungi in sandy dune soil could not only enrich soil functional microbial bank,but also accelerate litter decomposition,available nutrient input,and sandy land restoration in the Horqin Sandy Land.
2 Materials and methods
2.1 Study area
This study was conducted at the Naiman Desertification Research Station of the Chinese Academy of Sciences,which is located in Naiman County in the southwestern part of the Horqin Sandy Land,Inner Mongolia,China(42°55'N,120°42'E;360 m a.s.l.).The climate is temperate,semi-arid,continental monsoonal,receiving an average annual precipitation of 360 mm,more than 75% of which occurs in the growing season from June to September.Annual mean open-pan evaporation is 1,935 mm.Annual mean wind velocity ranges from 3.2–4.1 m/s,and the dominant wind is southwest to south in summer and autumn,and northwest in winter and spring.The zonal soil is sandy chestnut,which is sandy in texture,light yellow in color,and loose in structure,making it vulnerable to wind erosion(Zhaoet al.,2010).The landscape is characterized by mobile,semi-fixed,and fixed sand dunes,and interdune lowlands.Caragana microphyllaLam.,Artemisia halodendronTurcz.ex Besser,Melissitus ruthenicus(L.)Latsch.,Cleistogenes squarrosa(Trin.)Keng,Lespedeza davurica(Laxm.)Schindl.,Setaria viridis(L.)P.Beauv.,Pennisetum centrasiaticumTzvel.,andAgriophyllum squarrosum(L.)Moq.are characteristic plant species in the region.Afforestation to combat desertification has been performed with varying success,and the dominant tree and shrub species used areUlmus pumilaL.,Pinus sylvestrisL.,Populus simoniiCarr.,andC.microphyllaLam.(Zhaoet al.,2003).
2.2 Experimental design
2.2.1 Soil collection
We had chosen mobile,fixed,shrub,and afforestation dunes to collect the soil from the upper layer(0–20 cm).Five quadrats(5m×5m)were randomly selected as sampling sites in each type of dune,and five replications of soil cores were taken and mixed as a pooled sample in each quadrat.Every pooled sample was sieved(<2 mm)to remove rocks and plant material,and stored separately in sealed plastic bags at 4 °C for isolation of cellulose decomposing fungi.
2.2.2 Media
1)CMC medium.CMC-Na:15 g;yeast extract:1 g;KH2PO4:1 g;MgSO4·7H2O:0.5 g;NaCl:0.5 g;agar:14 g;H2O:1,000 mL;natural pH.
2)PDA medium.Potato:200 g;glucose:20 g;agar:20 g;H2O:1,000 mL;natural pH with penicillin.
3)Congo Red CMC medium.CMC-Na:20g;MgSO4·7H2O:0.5 g;KH2PO4:1 g;(NH4)2SO4:0.2 g;NaCl:0.5 g;Congo Red:0.2 g;agar:20 g;H2O:1,000 mL;natural pH.
4)Litter medium.KH2PO4:1 g;MgSO4·7H2O:0.3 g;NaNO3:2.5 g;FeCl3:0.01 g;CaCl2:0.1 g;NaCl:0.1 g;agar:18 g;litter powder:20 g;H2O:1,000 mL;pH:7.2.
5)Liquid medium.Peptone:2.6 g;yeast extract:1.3 g;litter powder:5 g;MgSO4·7H2O:0.8 g;KH2PO4:2 g;NaCl:0.1 g;H2O:1,000 mL.Take 100 mL for each bottle after stirring and sterilizing.
2.2.3 Screening procedures
Isolation:10 g of fresh soil were placed into 90 mL sterilized water and shaken until the soil suspension was uniform.1 mL of the supernate was placed into 9 mL sterilized water and allowed to stay for 10 min,to produce 10% and 1% soil suspensions.1 mL of 10% and 1% soil suspensions were placed into CMC medium with three replicates,respectively.The plates of CMC medium with the soil suspensions were incubated at 30 °C for 10–15 days.Colonies with different colors and larger rings in the plates were selected and numbered for purification.
Purification:Single strains were picked from the numbered colonies and transferred to PDA medium plates.The PDA medium plates with separated strains were incubated at 30 °C for 5 days for purification.Colonies without infection were chosen as selected fungi.
Pre-screening:Pure strains were placed into litter medium plates and incubated at 30 °C for 5 days.Colonies without infection were chosen as cellulose decomposing fungi.
Advanced-screening:Separated strains in the litter medium plates were scraped by vaccinating lancet and dipped it into Congo Red CMC medium plates to select the efficient cellulose decomposing fungi.Each strain was cloned in three plates and incubated at 30 °C for 10 days.Strains with larger rings and faster growth in the Congo Red CMC medium were screened as highly efficient cellulose decomposing fungi,and the selected fungi were stored in Congo Red CMC medium tubes for further DNA identification.
2.2.4 rDNA-ITS molecular identification
A small amount of fungi mycelium was selected to extract DNA(Zhanget al.,2008).Universal primers of ITS1(5'-TCCGTAGGTGAACCTGCGG-3')and ITS4(5'-TCCTCCGCTTATTGATATGC-3')were used for PCR.The PCR products sequence were tested in Nuosai Gene.The Blast alignment analysis was used to compare DNA sequence with GenBank database in NCBI,and MEGA 5 was used to build phylogenetic trees based on the DNA sequence,in order to determine the species of selected fungus.
2.2.5 CMC enzyme activity
Carboxylmethyl cellulose(CMC)enzyme activity(Fanget al.,2007;Bayeret al.,2013):The selected strain was incubated in a liquid medium by shaking(150 r/min)at 30 °C for 7 days.0.5 mL of supernate was placed into a tube after centrifuging the liquid culture(4,000 r/min,15 min),adding 1.5 mL citrate buffer(0.05 mol/L,pH=4.4,containing 0.5%CMC-Na)into the tube.The tube was placed in a water-bath at 50 °C for 30 min.1 mL DNS was added into the tube immediately after the water-bath,then treated the tube at 100 °C for 5 min.Water was added into the tube to a constant volume of 5 mL when the tube cooled down,and determine the absorbance at 540 nm in a spectrophotometer.The glucose content was calculated based on the standard curve.
CMC enzyme activityX = m/(V•t)•n
wheremrepresents glucose content,Vrepresents supernate volume(0.5 mL),trepresents reaction time(30 min)andnrepresents dilution ratio(5/0.5).
2.2.6 Decomposition ability
Cloth decomposition was used to test fungi decomposition ability(Yao and Huang,2006).Place a small piece of mycelium from the selected single strain into a liquid medium bottle,and incubate the fungi for 24 h.Then place five starch-free cloth pieces into each bottle,and keep the incubation in a shaking table(150 r/min)at 30 °C for 30 days.Determine the cloth weight loss on the 5th,10th,and 30th day.Only liquid medium without any strain was used as control(CK).
Cloth decomposition rate =(m0-mi)/m0×100%wherem0is the original cloth weight,whilemiis the cloth weight at theith day.
2.3 Data analysis
Data were analyzed and described by Microsoft Excel,SPSS 17.0 and Origin 8.0 for Windows.Values were presented as mean ± SE,and significant differences among treatment values were calculated by one-way analysis of variance(ANOVA).Least significant difference(LSD)tests were performed to evaluate differences among individual treatments.
3 Results
3.1 Screening
Table 1 shows that 233 cellulose decomposing fungi were detected in the CMC medium plates,and 31 were purified from the isolation due to different colony characteristics(such as color and shape)of the isolated strains.After pre-screening in litter powder medium,17 strains which grew faster without infection were selected.There were 1,3,7 and 6 strains originally from mobile,fixed,shrub and afforestation dunes,respectively.They are labeled as MD1,FD1,FD2,FD3,SD1,SD2,SD3,SD4,SD5,SD6,SD7,AD1,AD2,AD3,AD4,AD5 and AD6.Advanced screening shows that four strains(FD2,AD3,AD4 and SD4)grew better in Congo Red CMC medium as high-efficient cellulose decomposing fungi,and they are numbered as NM3-1,NM3-2,NM3-3 and NM3-4.The colony characteristics of the selected four high-efficient cellulose decomposing fungi are presented in Figure 1.
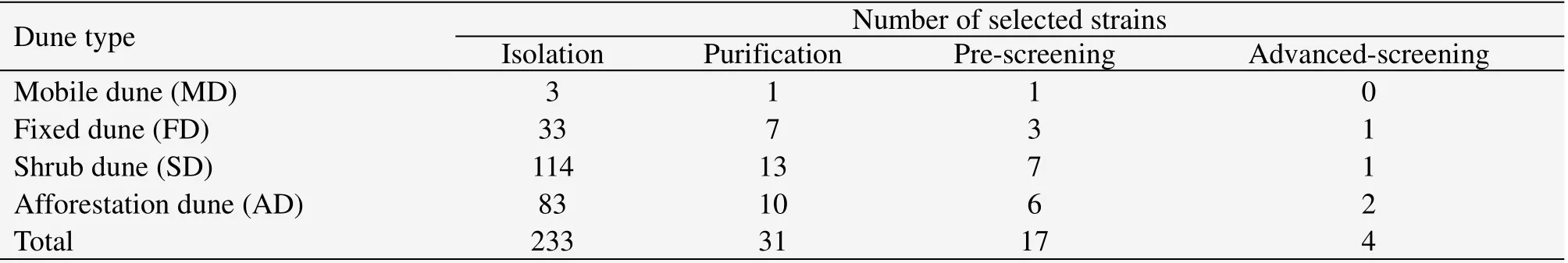
Table 1 Numbers of selected strains during screening procedure
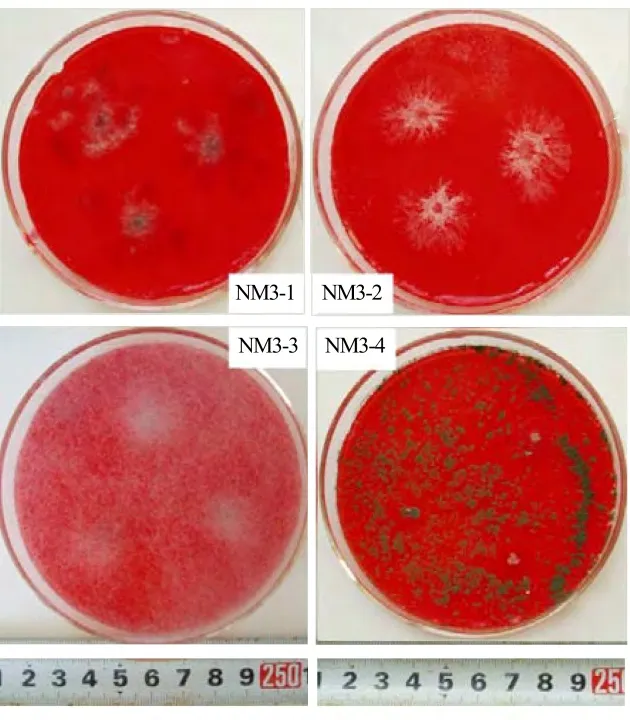
Figure 1 Colony characteristics of highly efficient cellulose decomposing fungi in Congo Red cellulose medium
3.2 Identification
The results of DNA sequencing analysis are presented in Table 2:the length of extracted DNA sequences of the four selected strains extended from 746–872 bp,and the G+C content reached from 45%–60%.Blast alignment of the four DNA sequences in Genbank database shows that they are mostly associated with FN907954.1,JX144267.1,JN222393.1 and JX173856.1,respectively.They also show high nucleoride homology with known fungal sequences in NCBI Genbank database(≥99%).Landeweertet al.(2003)explained that the strains are in the same family if the similarity of tested DNA sequence with known fungal sequence in Genbank database is 90%–95%;the strains are in the same genera if the similarity is 95%–99%;and the strains are the same species if the similarity is≥99%.The phylogenies of the DNA clones were analyzed.The phylogenetic trees of the selected and related strains are presented in Figures 2–5.According to DNA sequencing analysis,the four selected cellulose decomposing fungi are identified asAsperigillus calidoustus,Fusarium oxysporum,F.solaniandHypocrea lixii.The Molecular phylogenetic tree of the four selected strains are presented in Figure 6.

Table 2 Identification of DNA sequences and their taxonomic affiliations

Figure 2 Phylogenetic tree of NM3-1 and related strains
Figure 3 Phylogenetic tree of NM3-2 and related strains

Figure 4 Phylogenetic tree of NM3-3 and related strains

Figure 5 Phylogenetic tree of NM3-4 and related strains
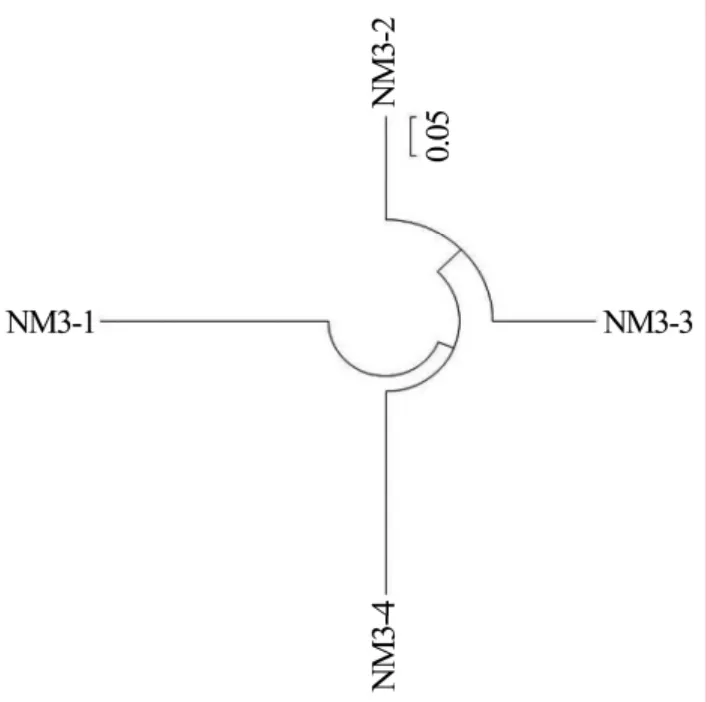
Figure 6 Molecular phylogenetic tree of the four selected strains
3.3 CMC enzyme activity
CMC enzyme activity of the four selected highefficient cellulose decomposing fungi show that the highest CMC enzyme activity was produced by NM3-4,which reached 0.196 mg/(mL·min),and the following were produced by NM3-3,NM3-1 and NM3-2,respectively.The CMC enzyme activity of NM3-4 was significantly higher than that of NM3-2(Table 3).
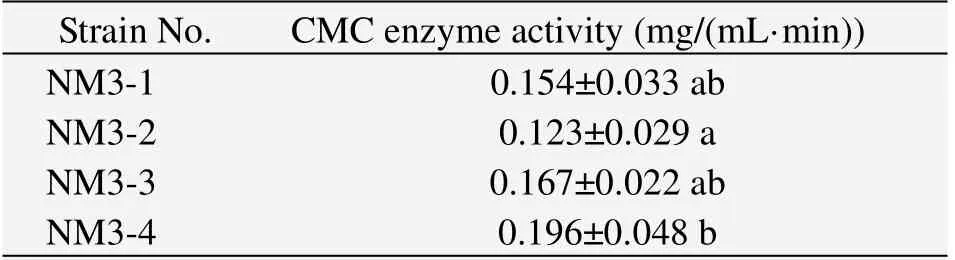
Table 3 CMC enzyme activity of the four selected strains
3.4 Decomposition ability
The results shows that cloth decomposition rate affected by the four selected fungi were significantly higher than that without a strain(CK).Cloth decomposition ability of the four selected fungi shows no significant difference.The highest cloth decomposition rate was 17.98% in 30 days affected by NM3-4.Cloth residual was reduced with the decomposition procedure,and decomposition rate appeared significantly higher at the 30th day than at the 5th and 10th day.
4 Discussion
There are numerous cellulose decomposers in the dune soil of Horqin Sandy Land.The 233 colonies were detected and 31 were isolated for purification.Four high-efficient cellulose decomposing fungi were isolated and screened in dune soil of Horqin Sandy Land,and they were identified asAsperigillus calidoustus,Fusarium oxysporum,F.solaniandHypocrea lixiiby rDNA-ITS sequencing analysis.Asperigillus,FusariumandHypocreaare widely present in the soil environment,and many species have high ability for litter decomposition(Abdel-Hafez,1982;Españaet al.,2011;Liuet al.,2011;Panagiotouet al.,2011;Mazaet al.,2014).
The four high-efficient cellulose decomposing fungi,screened from dune soil of Horqin Sandy Land,included oneAsperigillus(from fixed dune),twoFusarium(from afforestation dune)and oneHypocrea(from shrub dune).The species of high-efficient cellulose decomposing fungi are mostly different in different habitats,because chemical and biological litter properties are different in different dune habitats.Litter is mostly grass-derived in fixed dune,tree-derived in afforestation dune,and shrub-derived in shrub dune,and different decomposing fungi prefer different litter.More cellulose decomposing fungi were isolated and pre-screened in shrub dune than in afforestation dune,but we found more high-efficient cellulose decomposing fungi in afforestation dune(twoFusarium)than that in shrub dune(oneHypocrea).This may be due to the fact that litter in afforestation dune is mostly tree-derived and more difficult to decompose,thus more decomposers developed the ability for high-efficient decomposition ability.We detected several cellulose decomposing fungi in mobile dune,but none of them were high-efficient.This is mostly due to little litter input in mobile dune and cellulose decomposing fungi cannot survive in low nutrient environment.
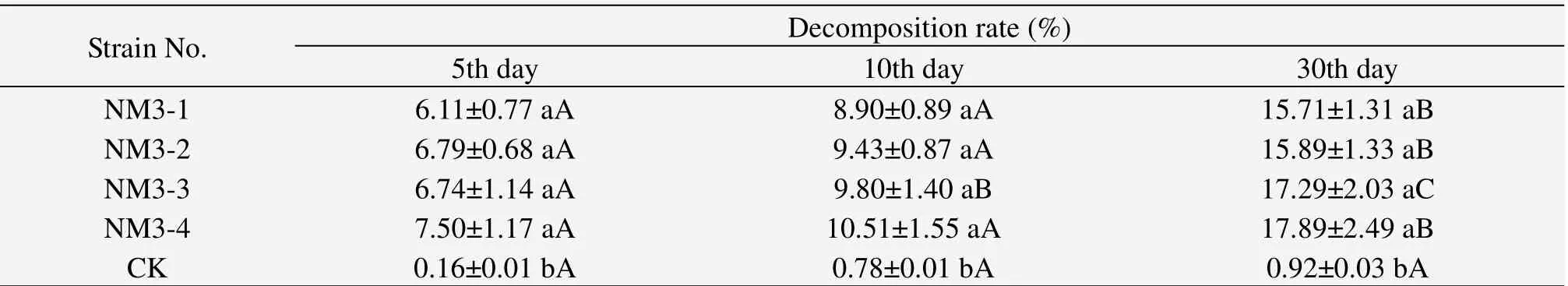
Table 4 Cloth decomposition rate
Litter is decomposed very slowly in natural sandy dunes(Quet al.,2011).The screened decomposing fungi could produce high amount of cellulase,and they multiplied rapidly in a laboratory environment.Thus,there is the possibility of accelerating litter decomposition in natural sandy dunes.If high-efficient cellulose decomposing fungi were added in sandy soil,soil microbial environment may change to some extent,and soil CMC enzyme activity could be higher as long as the screened decomposing fungi adapted to the environment.In this case,litter decomposition will be accelerated,adding more nutrient input and faster material cycling in sandy dune ecosystem.
5 Conclusion
Thirty-one stains were isolated to select efficient cellulose decomposers,and four efficient cellulose decomposing fungi were screened in dune soil of Horqin Sandy Land.They were identified asAsperigillus calidoustus,Fusarium oxysporum,F.solaniandHypocrea lixiiby rDNA-ITS molecular biological methods.Screening of efficient cellulose decomposers can not only increase dune soil functional microbe bank,but also accelerate litter decomposition and available nutrient input in Horqin Sandy Land.
This paper was financially supported by the National Science and Technology Support Program(2011BAC07B02),National Natural Science Foundation of China(41401620 and 41171414)and the Key Laboratory of Desert and Desertification Foundation(KLDD-2014-010)from Cold and Arid Regions Environmental and Engineering Research Institute,CAS.We express our sincere thanks to the anonymous reviewers for their valuable comments and suggestions on this manuscript,and to all the members of Naiman Desertification Research Station,CAS,for their help in field and laboratory work.
Abdel-Hafez S,1982.Cellulose-decomposing fungi of desert soils in Saudi Arabia.Mycopathologia,78:73–78.DOI:10.1007/BF00442629.
Bisaria VS,Ghose TK,1981.Biodegradation of cellulosic materials:substrates,microorganisms,enzymes and products.Enzyme and Microbial Technology,3:90–104.DOI:10.1016/0141-0229(81)90066-1.
Bayer EA,Shoham Y,Lamed R,2013,et al.,2013.Lignocellulose-decomposing bacteria and their enzyme systems.In:Rosenberg E,DeLong EF,Lory S,et al.(eds.).The Prokaryotes.Springer Berlin Heidelberg,pp.215–266.DOI:10.1007/978-3-642-30141-4_67.
Chapin III FS,Chapin MC,Matson PA,et al.,2011.Principles of Terrestrial Ecosystem Ecology.Springer.
España M,Rasche F,Kandeler E,et al.,2011.Assessing the effect of organic residue quality on active decomposing fungi in a tropical Vertisol using15N-DNA stable isotope probing.Fungal Ecology,4:115–119.DOI:10.1016/j.funeco.2010.09.005.
Fan C,Li SJ,Li CL,et al.,2012.Isolation,identification and cellulase production of a cellulolytic bacterium from intestines of giant panda.Acta Microbiologica Sinica,52:1113–1121.
Fang XT,Chen H,Zhao XF,et al.,2007.Determination of enzyme activity of straw cellulose-decomposing microorganisms.Letters in Biotechnology,18:628–630.
Hou Y,Wang QF,Chen Q,et al.,2013.Screening of high efficient cellulose-decomposing microorganisms.Research Journal of Chemistry and Environment,17:2–7.
Kamolmanit B,Vityakon P,Kaewpradit W,et al.,2013.Soil fungal communities and enzyme activities in a sandy,highly weathered tropical soil treated with biochemically contrasting organic inputs.Biology and Fertility of Soils,49:905–917.DOI:10.1007/s00374-013-0785-7.
Kubicek CP,Messner R,Gruber F,et al.,1993.TheTrichodermacellulase regulatory puzzle:From the interior life of a secretory fungus.Enzyme and Microbial Technology,15:90–99.DOI:10.1016/0141-0229(93)90030-6.
Landeweert R,Leeflang P,Kuyper TW,et al.,2003.Molecular identification of ectomycorrhizal mycelium in soil horizons.Applied and Environmental Microbiology,69:327–333.DOI:10.1128/AEM.69.1.327-333.2003.
Liu DY,Zhang RF,Yang XM,et al.,2011.Expression,purification and characterization of two thermostable endoglucanases cloned from a lignocellulosic decomposing fungiAspergillus fumigatusZ5 isolated from compost.Protein Expression and Purification,79:176–186.DOI:10.1016/j.pep.2011.06.008.
Lu GX,Chen XR,Yang CD,et al.,2011.Identification of cellulose decomposition fungi strain F1 and decomposition activity to two kinds of lawn grass litter.Acta Prataculturae Sinica,20:170–179.DOI:10.11686/cyxb20110622.
Lynd LR,Weimer PJ,van Zyl WH,et al.,2002.Microbial cellulose utilization:fundamentals and biotechnology.Microbiology and Molecular Biology Reviews,66:506–577.DOI:10.1128/MMBR.66.3.506-577.2002.
Maza M,Pajot HF,Amoroso MJ,et al.,2014.Post-harvest sugarcane residue degradation by autochthonous fungi.International Biodeterioration &Biodegradation,87:18–25.DOI:10.1016/j.ibiod.2013.10.020.
Niu JL,Li GX,Cui ZJ,et al.,2005.Construction and function of a high-efficient complex microbial system to degrade cellulose and lindane in compost.Environmental Science,26:186–190.DOI:10.3321/j.issn:0250-3301.2005.04.037.
Panagiotou G,Kekos D,Macris BJ,et al.,2003.Production of cellulolytic and xylanolytic enzymes byFusarium oxysporumgrown on corn stover in solid state fermentation.Industrial Crops and Products,18:37–45.DOI:10.1016/S0926-6690(03)00018-9.
Panagiotou G,Topakas E,Moukouli M,et al.,2011.Studying the ability ofFusarium oxysporumand recombinantSaccharomyces cerevisiaeto efficiently cooperate in decomposition and ethanolic fermentation of wheat straw.Biomass and Bioenergy,35:3727–3732.DOI:10.1016/j.biombioe.2011.05.005.
Qu H,Zhao XY,Zhao HL,et al.,2011.Litter decomposition rates in Horqin Sandy Land,Northern China:Effects of habitat and litter quality.Fresenius Environmental Bulletin,20:3304–3312.
Regnell O,Elert M,Glund LOH,et al.,2014.Linking cellulose fiber sediment methyl mercury levels to organic matter decay and major element composition.Ambio,43:878–890.DOI:10.1007/s13280-013-0487-2.
Schwarz W,2001.The cellulosome and cellulose degradation by anaerobic bacteria.Applied Microbiology and Biotechnology,56:634–649.DOI:10.1007/s002530100710.
Štursová M,Žifčáková L,Leigh MB,et al.,2012.Cellulose utilization in forest litter and soil:identification of bacterial and fungal decomposers.Fems Microbiology Ecology,80:735–746.DOI:10.1111/j.1574-6941.2012.01343.x.
Wang SK,Zhao XY,Zhang TH,et al.,2013.Afforestation effects on soil microbial abundance,microbial biomass carbon and enzyme activity in dunes of Horqin Sandy Land,northeastern China.Sciences in Cold and Arid Regions,5:184–190.DOI:10.3724/SP.J.1226.2013.00184.
Wen ZY,Liao W,Chen SL,2005.Production of cellulase byTrichoderma reeseifrom dairy manure.Bioresource Technology,96:491–499.DOI:10.1016/j.biortech.2004.05.021.
Yao HY,Huang CY,2006.Soil Microbial Ecology and Their Research Approaches.Beijing:Science Press.
Zhang YH,Wei DL,Xing LJ,et al.,2008.A modified method for isolating DNA from Fungus.Microbiology China,35:466–469.
Zhao HL,Zhao XY,Zhang TH,et al.,2003.Desertification Processes and Its Restoration Mechanisms in the Horqin Sand Land.Beijing:Ocean Press.
Zhao XY,Luo YY,Wang SK,et al.,2010.Is desertification reversion sustainable in Northern China?––A case study in Naiman County,part of typical agro-pastoral transitional zone in Inner-Mongolia,China.Global Environmental Research,14:63–70.
杂志排行
Sciences in Cold and Arid Regions的其它文章
- Toward sustainable desertification reversion:A case study in Horqin Sandy Land of northern China
- Land desertification and restoration in Middle East and North Africa(MENA)region
- Effects of land-use changes on organic carbon in bulk soil and associated physical fractions in China's Horqin Sandy Grassland
- Adaptation to climate change in desertified lands of the marginal regions in Egypt through sustainable crop and livestock diversification systems
- A mechanism for the origin and development of the large-scale dunefield on the right flank of the lower reach of Laoha River,Northeast China
- Seasonal change mediates the shift between resource and pollen limitation in Hedysarum scoparium(Fabaceae)
