Aberrant methylation frequency of TNFRSF10C promoter in pancreatic cancer cell lines
2011-07-07HuiHuaCaiYueMingSunYiMiaoWenTaoGaoQuanPengJieYaoandHanLinZhao
Hui-Hua Cai, Yue-Ming Sun, Yi Miao, Wen-Tao Gao, Quan Peng, Jie Yao and Han-Lin Zhao
Nanjing, China
Aberrant methylation frequency of TNFRSF10C promoter in pancreatic cancer cell lines
Hui-Hua Cai, Yue-Ming Sun, Yi Miao, Wen-Tao Gao, Quan Peng, Jie Yao and Han-Lin Zhao
Nanjing, China
BACKGROUND:A growing body of evidence suggests that many tumors are initiated by both epigenetic abnormalities and gene mutations, which promote tumor progression. Epigenetic abnormalities include changes in DNA methylation and in the modification of histones. This study aimed to assess the status of methylation in the CpG island (CGI) of the tumor necrosis factor receptor superfamily member 10c (TNFRSF10C) with combined bisulfite restriction analysis (COBRA) and to evaluate its role in the progression of pancreatic cancer (PC).
METHODS:The methylation status of four PC cell lines was assessed using COBRA and/or bisulfite genomic sequencing (BGS). Changes in methylation and TNFRSF10C expression in PC cell lines before and after treatment with 5-aza-2'-deoxycytidine (5-aza-dC) and/or trichostatin A (TSA) were assessed by BGS and real-time RT-PCR. Apoptosis in the four cell lines was tested by flow cytometry (FCM) and TUNEL assay.
RESULTS:The methylation status of the TNFRSF10C promoter was assessed in PC cells (BxPC-3: 68.84±8.71%; CFPAC-1: 0; PANC-1: 96.77±4.57%; SW1990: 54.97±7.33%) with the COBRA assay, which was confirmed by the results of BGS. After treatment with 5-aza-dC and/or TSA, apoptosis was induced in PC cells to different degrees, and the levels of TNFRSF10C transcriptional expression in the PC cell lines (except CFPAC-1) increased markedly after 5-aza-dC treatment.
CONCLUSIONS:A high frequency of CGI methylation in the TNFRSF10C promoter results in inactivation of the gene and enhancement of tumor growth in most PC celllines (except CFPAC-1). Inactivation of TNFRSF10C by CGI hypermethylation can play an important role in PC progression and be potentially useful as a diagnostic marker and a new therapeutic approach for PC.
(Hepatobiliary Pancreat Dis Int 2011; 10: 95-100)
methylation; CpG island; pancreatic cancer; TNFRSF10C
Introduction
Pancreatic cancer (PC) is a malignant tumor associated with a significant degree of morbidity and mortality.[1]It is highly invasive and responds poorly to conventional treatments. Like other tumor types, PC arises from a complex and poorly understood sequence of genetic and epigenetic alterations. Aberrant hypermethylation of CpG islands (CGIs) in gene promoters and histone deacetylation are the main epigenetic changes that lead to gene silencing, but this has not been thoroughly investigated in PC cell lines. Elucidating the mechanisms of these regulatory events is likely to enhance our understanding of this disease and the identification of new treatment targets.
The tumor necrosis factor receptor superfamily member 10c (TNFRSF10C) is one of several TNF-related, apoptosis-inducing ligand (TRAIL)-like decoy receptors. It is a glycosyl phosphatidylinositol-linked membrane molecule that lacks a cytoplasmic region and is located on 8p22-p21.[2]TNFRSF10C is one of the most frequently deleted loci in cancers of the colon,[3]lung,[4]prostate,[5,6]breast[7,8]and bladder.[9]Hypermethylation, specifically of the CGI in the TNFRSF10C promoter, has been reported in neuroblastoma (21%),[10]primary breast cancer (48%), primary lung cancer (37%), malignant mesothelioma (43%),[8]ependymoma (50%), and choroid plexus papilloma (50%).[11]Furthermore, a recent study by Hornstein et al[12]reported a significant decrease in expression of TNFRSF10C in the M0 stage of prostate carcinoma tissue compared to benign prostate tissue.These data suggest that TNFRSF10C is a tumor suppressor gene of prostate carcinoma. However, hypermethylation of the TNFRSF10C promoter in PC has not previously been investigated.
In the present study, we determined the frequency of hypermethylation in the TNFRSF10C promoter in four PC cell lines (BxPC-3, CFPAC-1, PANC-1, and SW1990) by using combined bisulfite restriction analysis (COBRA) and/or bisulfite genomic sequencing (BGS). Changes in methylation status and gene expression of TNFRSF10C before and after treatment with 5-aza-2'-deoxycytidine (5-aza-dC) or trichostatin A (TSA) were also evaluated in these PC cell lines.
Methods
Cell culture and drug treatment
The PC cell lines (BxPC-3, CFPAC-1, PANC-1, and SW1990) were cultured in DMEM (Hyclone, Logan, UT), and all cells were maintained at 37 ℃ and 5% CO2in a humidified incubator. Treatments of cell lines were: 100 nmol/L TSA (Sigma, St. Louis, MO) for 24 hours, 5 μmol/L 5-aza-dC (Sigma, St. Louis, MO) for 72 hours (with fresh drug added every 24 hours), and 5 μmol/L 5-aza-dC for 72 hours followed by 24 hours with 100 nmol/L TSA.[13]The control group was untreated. Apoptosis before and after drug treatment was assayed using flow cytometry (FCM) and TUNEL assay.
Isolation of RNA and genomic DNA
RNA from cells was isolated using Trizol according to the manufacturer's instructions. Recovered RNA was further treated with DNaseiand re-purified using a Micro-to-Midi total RNA purification system (Invitrogen, Carlsbad, CA, USA).
Genomic DNA from cultured cells was isolated from the inter-organic phase of a Trizol-chloroform mixture, then combined with 0.5 ml of back extraction buffer (4 mol/L guanidine thiocyanate, 50 mmol/L sodium citrate, 1 mol/L Tris, pH 8.0). After vigorous mixing by inversion, phase separation was achieved by centrifugation at 12 000×g for 30 minutes. DNA was precipitated from the upper aqueous phase using isopropanol and further purified using Gentra Puregene reagent (Qiagen, Hilden, Germany) with proteinase K treatment.[14]
Bisulfite modification
DNA (4 μg) was denatured with 0.2 mol/L NaOH for 20 minutes at 37 ℃ and treated with sodium bisulfite (Sigma, St Louis, MO) for 16 hours at 50 ℃. Modified DNA was purified using the Wizard DNA Clean-up system (Promega, Madison, WI) and diluted in 50 μl nuclease-free water (NFH2O). Modification was completed by incubation with NaOH (0.3 mol/L final concentration) for 5 minutes at room temperature. DNA was precipitated with ethanol and resuspended in 60 μl NFH2O. DNA from SW1990 cells was treated with CpG methyltransferase (M.SssI) (NEB, Ipswich, MA) according to the manufacturer's protocol and modified by sodium bisulfite for a positive control.
BGS
TNFRSF10C methyl primers were designed using Methyl-Primer-Express software v1.0 (Applied Biosystems, Foster City, CA) to amplify bisulfite-modified DNA. The forward and reverse primer sequences were: 5'-GGA TCC CCA AGA CCC TAA AG-3' and 5'-TGT TGG AAG CGT TGG TGT AA-3', respectively, and a 215-bp product amplified contained 10 CpG dinucleotides. PCR samples contained 36.3 μl NFH2O, 5 μl 10×buffer (Mg2+), 2.5 μl 10 mmol/L dNTP, 2 μl of each primer, 2 μl bisulfite-treated DNA, and 0.2 μl FastTaqTMpolymerase (Qiagen, Hilden, Germany). Amplification was performed with the following cycles: (a) 95 ℃ for 5 minutes; (b) 2 cycles of 95 ℃for 45 seconds, 68 ℃ for 30 seconds, and 72 ℃ for 30 seconds; (c) 3 cycles of 95 ℃ for 45 seconds, 66 ℃ for 30 seconds, and 72 ℃ for 30 seconds; (d) 4 cycles of 95 ℃ for 45 seconds, 64 ℃ for 30 seconds, and 72 ℃for 30 seconds; (e) 5 cycles of 95 ℃ for 45 seconds, 62 ℃ for 30 seconds, and 72 ℃ for 30 seconds; (f) 35 cycles of 95 ℃ for 45 seconds, 60 ℃ for 30 seconds, and 72 ℃ for 30 seconds; with a final extension step for 7 minutes at 72 ℃. Amplified PCR products (5 μl) were separated by electrophoresis in 2% agarose gels and visualized by ethidium bromide staining. Using the Toyobo TA cloning kit (Toyobo, Osaka, Japan), 2 μl of PCR product was subcloned into the pCR4-TOPO vector and transformed into E. coli. Sequencing analysis was performed on 10-20 randomly-selected individual colonies and the data were compared with the UCSC genome reference sequence to assess the methylation status of each CpG site.
COBRA[15]
The restriction enzyme TaqI (NEB, Ipswich, MA) was included in COBRA assay samples as follows: 18 μl NFH2O, 2 μl 10×buffer, 10 μl PCR product, and 2 μl TaqI. Reaction conditions were (a) 65 ℃ for 16 hours and (b) 80 ℃ for 20 minutes. Digested PCR products were separated on an 8% polyacrylamide gel, visualized by ethidium bromide staining, and photographed usinga Bio-Rad gel documentation system. Quantitation was performed using a Molecular Dynamics phosphor imager.
Quantitative RT-PCR
Total RNA was purified from cultured cells before and after treatment with 5-aza-dC or TSA using the RNeasy kit (Qiagen, Hilden, Germany). cDNA was synthesized using SuperScript II reverse transcriptase and Oligo (dT) primers (Invitrogen, Carlsbad, CA) in a 20 μl reaction according to the manufacturer's protocol. For the PCR reactions, Platinum SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen, Carlsbad, CA) was used and reactions were assayed in triplicate using 1 μl of cDNA as a template in a 25 μl reaction. β-actin was used as the reference gene. Primers for TNFRSF10C were: 5'-TAC ACC AAC GCT TCC AAC-3' (forward) and 5'-AAG TGT AGC AGG TGC CCT-3' (reverse). TNFRSF10C PCR was performed with the following cycles: 95 ℃ for 10 minutes, 95 ℃ for 15 seconds, 63 ℃ for 32 seconds, and 72 ℃ for 30 seconds. A total of 40 cycles was performed and each reaction was run in duplicate. Negative controls were included in each experiment. Gene expression was calculated based on the yield of TNFRSF10C relative to β-actin.
Results
Apoptosis before and after drug treatment
The percentage of apoptotic cells after treatment with 5-aza-dC and/or TSA was greater than that of the control in FCM and the TUNEL assay (Fig. 1). Higher levels of apoptosis were measured in CFPAC-1, PANC-1, and SW1990 cells treated with 5-aza-dC than with TSA. In contrast, the cell line BxPC-3 exhibited higher levels of apoptosis after TSA treatment (Fig. 1B).
Methylation frequency of CGI in TNFRSF10C promoter
The methylation frequency of the CGI in the TNFRSF10C promoter was assessed in PC cell lines using the COBRA assay. Methylation was absent in CFPAC-1 cells, 96.77±4.57% in PANC-1 cells, 68.84±8.71% in BxPC-3 cells, and 54.97±7.33% in SW1990 cells (Fig. 2A, B).
At the same time, the DNAs of PANC-1 and CFPAC-1 were subjected to BGS. Ten CpG sites within the CGI of the TNFRSF10C promoter were detected and the percentages of methylation (PANC-1: 98%;CFPAC-1: 0%) were consistent with the results of the COBRA assay (Fig. 2C).
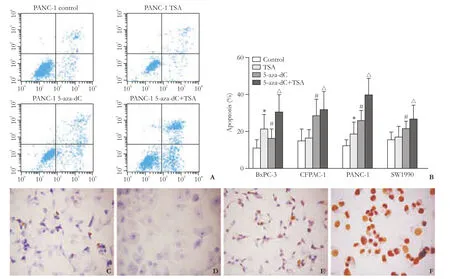
Fig. 1. Evaluation of apoptosis. A: Flow cytometric analysis of apoptosis exhibited by PANC-1 cells after treatment with 5-aza-dC and/or TSA compared to control cells. B: The different apoptosis of PC cell lines treated with the combination of 5-aza-dC and TSA. BxPC-3 was more strongly affected by histone modification than changes in methylation. *, #, △: P<0.05, compared with control group. C-F: Representative images of apoptosis detected by TUNEL assay (original magnification ×400); C: untreated PANC-1 cells; D: negative control; E: PANC-1 cells treated with TSA; F: PANC-1 cells treated with 5-aza-dC. Blue staining indicates the absence of apoptosis and brown staining indicates the presence of apoptosis.
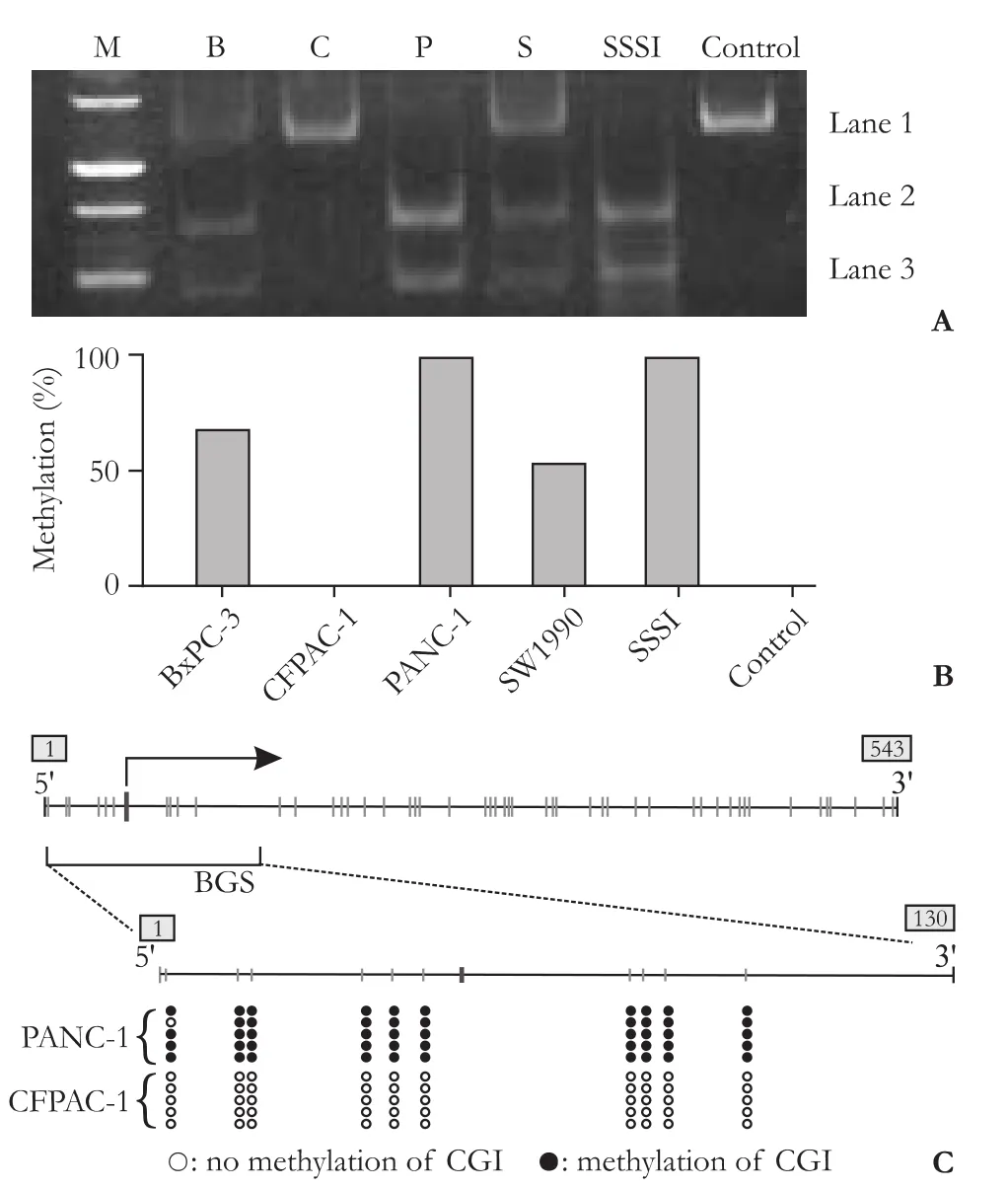
Fig. 2. Methylation frequency of CGI in TNFRSF10C promoter. A: COBRA results from four PC cell lines (M: marker; B: BxPC-3; C: CFPAC-1; P: PANC-1; S: SW1990; SSSI: DNA treated with CpG methyltransferase). B: The methylation rate of TNFRSF10C was calculated from the level of the gray lane (Fig. 2A) according to the following formula: methylation rate (%)=(lane 2+lane 3)/(lane 1+lane 2+lane 3). C: BGS of PANC-1 (98%), CFPAC-1 (0%).
Diversity of expression and changes in methylation before and after drug treatment
Real-time RT-PCR further demonstrated that demethylation was accompanied by a 8.28-fold (5-azadC) and a 5.75-fold (TSA) increase in transcriptional expression of TNFRSF10C in PANC-1, and a 2.86-fold (5-aza-dC) and a 1.70-fold (TSA) increase in SW1990 (Fig. 3A). Unlike the other cell lines, TNFRSF10C was found to be more strongly expressed after TSA treatment (3.64-fold) than 5-aza-dC treatment (2.98-fold) in BxPC-3 cells, which was paralleled by the FCM results (Fig. 1B). These data suggest that silencing of the TNFRSF10C gene is achieved through promoter methylation.
After 5-aza-dC or TSA treatment, BGS was used to re-assess the methylation status of TNFRSF10C in PANC-1 cells. The methylation rate in the untreated group was 98%, in the TSA group 70%, and in the 5-aza-dC group 40% (Fig. 3B).
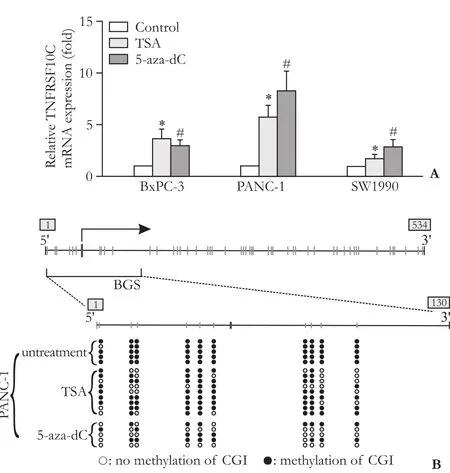
Fig. 3. Reduced methylation and reactivation of TNFRSF10C expression in PC cell lines after treatment with 5-aza-dC or TSA. A: Quantitative RT-PCR of TNFRSF10C mRNA after treatment of PC cell lines (CFPAC-1 is omitted) with 5-aza-dC or TSA. Untreated cells were used as control in each case. *, #: P<0.05, compared with the control group. B: BGS of the CGI of TNFRSF10C after treatment with 5-aza-dC or TSA in PANC-1 (untreated: 98%; TSA: 70%; 5-aza-dC: 40%).
Discussion
COBRA is a sensitive and quantitative assay to determine DNA methylation levels at specific gene loci in small amounts of genomic DNA. Restriction enzyme digestion is used to reveal methylation-dependent sequence differences in the PCR products of sodium bisulfitetreated DNA, and the results are verified by BGS. We evaluated the aberrant methylation frequency of TNFRSF10C in PC cell lines with the COBRA assay. Using a combined approach of pharmacologic inhibition of epigenetic modifications and gene expression assays, genes subject to epigenetic silencing in PC cell lines were identified. This strategy is also used to identify the targets of epigenetic silencing in other tumor types.[7,16-18]
TNFRSF10C lacks both a cytoplasmic and a transmembrane domain, leaving it unable to participate in apoptotic signaling. Since a direct correlation between TNFRSF10C expression and TRAIL sensitivity has not been demonstrated,[19-21]the role of TNFRSF10C in PC is still unknown.
In our study, hypermethylation of the TNFRSF10C promoter was found in all PC cell lines except CFPAC-1, but the mechanism is unclear. The results demonstrated a high prevalence of TNFRSF10C promoter CGI hyper-methylation in most PC cell lines. And the results of real-time RT-PCR showed low transcription in PC cells because of hypermethylation. After treatment with 5-aza-dC or TSA, we found that the methylation rate of the TNFRSF10C promoter CGI was cut from 98% to 40% (5-aza-dC) and to 70% (TSA) in PANC-1, and the transcriptional expression was enhanced by 8.28-fold (5-aza-dC) and 5.75-fold (TSA). Meanwhile, the apoptotic rate increased markedly. Thus gene silencing of TNFRSF10C is mostly due to aberrant hypermethylation, which plays an important role in the progression of PC. At the same time, 5-aza-dC and TSA have synergistic effects on demethylation to a certain extent, but the mechanism of demethylation by TSA is not clear. In the present study COBRA and BGS showed no methylation of the TNFRSF10C promoter in CFPAC-1 while apoptosis was still induced after 5-aza-dC and/or TSA treatment. The possible reason is that other anti-oncogenes were re-activated because there was no specific demethylation effect of 5-aza-dC. Both apoptosis and transcription of BxPC-3 treated with TSA were greater than that with 5-aza-dC, indicating that histone deacetylation mainly contributes to the progression in BxPC-3. On the contrary, hypermethylation plays a key role in progression in PANC-1 and SW1990. All the above suggest that different mechanisms result in tumorigenesis and progression in different PC cell lines.
With the increasing incidence of morbidity and high mortality, PC responds poorly to conventional treatments. Evaluation of aberrant CGI methylation frequency in the TNFRSF10C promoter may be used as a biomarker of PC. The methylation status of the TNFRSF10C promoter should be investigated in clinical PC specimens before it is used as a new therapeutic approach for PC.
Funding:The study was supported by a grant from the National Natural Science Foundation of China (30471691).
Ethical approval:Not needed.
Contributors:MY proposed the study. CHH and SYM carried out the study, and GWT, PQ and YJ collected and summarized the data. CHH wrote the first draft, which was corrected by MY and ZHL. All authors contributed to the intellectual context and approved the final version. MY is the guarantor.
Competing interest:No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
1 Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin 2008;58:71-96.
2 Ashkenazi A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer 2002;2: 420-430.
3 Van Geelen CM, de Vries EG, de Jong S. Lessons from TRAIL-resistance mechanisms in colorectal cancer cells: paving the road to patient-tailored therapy. Drug Resist Updat 2004;7: 345-358.
4 Tessema M, Yu YY, Stidley CA, Machida EO, Schuebel KE, Baylin SB, et al. Concomitant promoter methylation of multiple genes in lung adenocarcinomas from current, former and never smokers. Carcinogenesis 2009;30:1132-1138.
5 Chaib H, MacDonald JW, Vessella RL, Washburn JG, Quinn JE, Odman A, et al. Haploinsufficiency and reduced expression of genes localized to the 8p chromosomal region in human prostate tumors. Genes Chromosomes Cancer 2003;37:306-313.
6 Cheng Y, Kim JW, Liu W, Dunn TA, Luo J, Loza MJ, et al. Genetic and epigenetic inactivation of TNFRSF10C in human prostate cancer. Prostate 2009;69:327-335.
7 Wu Y, Alvarez M, Slamon DJ, Koeffler P, Vadgama JV. Caspase 8 and maspin are downregulated in breast cancer cells due to CpG site promoter methylation. BMC Cancer 2010;10: 32.
8 Shivapurkar N, Toyooka S, Toyooka KO, Reddy J, Miyajima K, Suzuki M, et al. Aberrant methylation of trail decoy receptor genes is frequent in multiple tumor types. Int J Cancer 2004; 109:786-792.
2.科研成果转化过程特征。科研成果转化在不同的阶段表现出不同的性质特点,有些是某一时点上的瞬间转化,有些是一个比较缓慢的过程。不同阶段不同形式的转化,都需要借助一定的条件来完成,研究的任务是找到这些条件,促成转化过程的完成。如表3所示:
9 Adams J, Williams SV, Aveyard JS, Knowles MA. Loss of heterozygosity analysis and DNA copy number measurement on 8p in bladder cancer reveals two mechanisms of allelic loss. Cancer Res 2005;65:66-75.
10 van Noesel MM, van Bezouw S, Salomons GS, Voute PA, Pieters R, Baylin SB, et al. Tumor-specific down-regulation of the tumor necrosis factor-related apoptosis-inducing ligand decoy receptors DcR1 and DcR2 is associated with dense promoter hypermethylation. Cancer Res 2002;62:2157-2161.
11 Michalowski MB, de Fraipont F, Michelland S, Entz-Werle N, Grill J, Pasquier B, et al. Methylation of RASSF1A and TRAIL pathway-related genes is frequent in childhood intracranial ependymomas and benign choroid plexus papilloma. Cancer Genet Cytogenet 2006;166:74-81.
12 Hornstein M, Hoffmann MJ, Alexa A, Yamanaka M, Müller M, Jung V, et al. Protein phosphatase and TRAIL receptor genes as new candidate tumor genes on chromosome 8p in prostate cancer. Cancer Genomics Proteomics 2008;5:123-136.
13 Kim TY, Zhong S, Fields CR, Kim JH, Robertson KD. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res 2006;66:7490-7501.
14 Warnecke PM, Stirzaker C, Song J, Grunau C, Melki JR, Clark SJ. Identification and resolution of artifacts in bisulfite sequencing. Methods 2002;27:101-107.
15 Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res 1997;25:2532-2534.
17 Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP, et al. A genomic screen for genes upregulated by demethylation and histone deacetylaseinhibition in human colorectal cancer. Nat Genet 2002;31:141-149.
18 Lai JC, Wu JY, Cheng YW, Yeh KT, Wu TC, Chen CY, et al. O6-Methylguanine-DNA methyltransferase hypermethylation modulated by 17beta-estradiol in lung cancer cells. Anticancer Res 2009;29:2535-2540.
19 Mahalingam D, Szegezdi E, Keane M, Jong S, Samali A. TRAIL receptor signalling and modulation: Are we on the right TRAIL? Cancer Treat Rev 2009;35:280-288.
20 Tarragona J, Llecha N, Santacana M, Lopez S, Gatius S, Llobet D, et al. DcR1 expression in endometrial carcinomas. Virchows Arch 2010;456:39-44.
21 Klugman SD, Gross SJ, Liang J, Livne K, Gross B, Khabele D, et al. Expression of Keratin 8 and TNF-Related Apoptosis-I Inducing Ligand (TRAIL) in Down Syndrome Placentas. Placenta 2008;29:382-384.
Accepted after revision September 2, 2010
Examine yourself every day. Correct the mistakes, if any, and guard against them, if not.
– (the Song Dynasty) Zhu Xi
May 20, 2010
Author Affiliations: Department of General Surgery, First Affiliated Hospital, Nanjing Medical University, Nanjing 210029, China (Cai HH, Sun YM, Miao Y, Gao WT, Peng Q, Yao J and Zhao HL); Department of General Surgery, Jiangsu Province Hospital of Traditional Chinese Medicine, Nanjing 210029, China (Cai HH)
Yi Miao, MD, PhD, Department of General Surgery, First Affiliated Hospital, Nanjing Medical University, Nanjing 210029, China (Tel: 86-25-83718836ext6508; Fax: 86-25-83781992; Email: miaoyi@njmu. edu.cn)
© 2011, Hepatobiliary Pancreat Dis Int. All rights reserved.
猜你喜欢
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Expression oflamino acid transport system 1 and analysis of iodine-123-methyltyrosine tumor uptake in a pancreatic xenotransplantation model using fused high-resolution-micro-SPECT-MRI
- Laparoscopic liver resection for benign and malignant liver tumors
- Monday blues of deceased-donor liver transplantation
- Hepatobiliary & Pancreatic Diseases International (HBPD INT)
- Roles of sulfonylurea receptor 1 and multidrug resistance protein 1 in modulating insulin secretion in human insulinoma
- Atypical focal nodular hyperplasia of the liver
