The OsBSK1-2-MAPK module regulates blast resistance in rice
2024-03-07ShengpingLiXinqunXingZhijunDioXiLingLuJingZhngZhiweiChenDingzhongTng
Shengping Li*, Xinqun Xing, Zhijun Dio,b, N Xi, Ling Lu, Jing Zhng,Zhiwei Chen,d, Dingzhong Tng,c,*
a State Key Laboratory of Ecological Control of Fujian-Taiwan Crop Pests, Key Laboratory of Ministry of Education for Genetics, Breeding and Multiple Utilization of Crops,Plant Immunity Center, Fujian Agriculture and Forestry University, Fuzhou 350002, Fujian, China
b College of Life Science, Fujian Agriculture and Forestry University, Fuzhou 350002, Fujian, China
c Ministerial and Provincial Joint Innovation Centre for Safety Production of Cross-Strait Crops, Fujian Agriculture and Forestry University, Fuzhou 350002, Fujian, China
d Fujian Provincial Key Laboratory of Crop Breeding by Design, Fujian Agriculture and Forestry University, Fuzhou 350002, Fujian, China
Keywords:Disease resistance Magnaporthe oryzae Oriza sativa Rice blast
ABSTRACT Receptor-like cytoplasmic kinase OsBSK1-2 was reported to play an important role in regulation of response to rice blast, but the signaling pathway remained unknown.In this study, we identified OsMAPKKK18 and previously uncharacterized MAPKKKs OsMAPKKK16 and OsMAPKKK19 that interact with OsBSK1-2.Expression of all three MAPKKKs was induced by Magnaporthe oryzae infection, and all three induced cell death when transiently expressed in Nicotiana benthamiana leaves.Knockout of OsMAPKKK16 and OsMAPKKK18 compromised blast resistance and overexpression of OsMAPKKK19 increased blast resistance, indicating that all three MAPKKKs are involved in regulation of rice blast response.Furthermore, both OsMAPKKK16 and OsMAPKKK19 interacted with and phosphorylated OsMKK4 and OsMKK5, and chitin-induced MAPK activation was suppressed in osmapkkk16 and osbsk1-2 mutants.OsMAPKKK18 was earlier reported to interact with and phosphorylate OsMKK4 and affect chitin-induced MAPK activation,suggesting that OsBSK1-2 is involved in regulation of immunity through multiple MAPK signaling pathways.Unlike BSK1 in Arabidopsis,OsBSK1-2 was not involved in response to avirulent M.oryzae strains.Taken together, our results revealed important roles of OsMAPKKK16/18/19 and a OsBSK1-2-OsMAPKKK16/18/19-OsMKK4/5 module in regulating response to rice blast.
1.Introduction
Exposed to the threat of various pathogens,plants have evolved a two-layered immune system to address infection: patterntriggered immunity (PTI) and effector-triggered immunity (ETI)[1].In PTI, pathogen/microbe-associated molecular patterns(PAMPs) and damage-associated molecular patterns (DAMPs) are recognized by pattern recognition receptors (PRRs), and immune responses are delivered to downstream signaling pathways by receptor-like cytoplasmic kinases (RLCKs) [2,3].RLCKs were reported to function as central components linking PRRs and downstream defence responses.For instance, in Arabidopsis,BOTRYTIS-INDUCED KINASE1(BIK1)receives immune signals from FLS2 and BAK1 and activates downstream signaling [4,5], and PBL27 acts as the link between cell surface chitin receptor CERK1 and the intracellular MAPK cascade [6].In rice, RLCK185 and RLCK176 are important defence-related proteins that function downstream of OsCERK1 in the peptidoglycan(PGN)and chitin signaling pathways [7,8].
BRASSINOSTEROID-SIGNALING KINASE1 (BSK1), a member of the RLCK XII subfamily, was previously shown to be a substrate of brassinosteroid insensitive 1 (BRI1) in Arabidopsis [9].BSK1 interacts with FLS2 and regulates flg22-induced immune responses[10].Further studies showed that BSK1 regulates plant immunity by interacting with and phosphorylating MAPKKK5 [11], and that BSK1 is involved in RECEPTOR-LIKE KINASE 902 (RLK902)-mediated resistance to bacteria [12].BSK1 was recently found to regulate powdery mildew resistance in Arabidopsis by affecting phosphorylation of MPK15 [13].OsBSK1-2, a BSK1 homologue in rice, functions as an important regulator in rice plant immunity,and silencing OsBSK1-2 decreased resistance to M.oryzae[14].Nevertheless, the mechanism by which OsBSK1-2 regulates disease resistance in rice remained unknown.
MAPK cascades are highly conserved signaling modules that are critical for immunity, stress responses and growth in plants[15–19].Activation of MAPK cascades is one of the earliest and most crucial events in plant defence response [17].Among MAPK cascades, MAPKKKs are the first responders, and they transmit defence signals from RLCKs to MAPKKs.For example, MAPKKK5 in Arabidopsis is activated by immunity signals from defencerelated RLCKs PBL27 and BSK1 and transduces signals to MAPKK4 and MAPKK5 by phosphorylation [6,11].Furthermore, three defence-related MAPKKKs, MEKK1, MAPKKK3 and MAPKKK5, link RLCK VII-4 subfamily members,which are central components that integrate signals from multiple PRRs, and downstream MAPK signal pathway components [20].It was reported that OsMAPKKK18 and OsMAPKKKε both deliver the chitin-triggered immunity signals from an OsCERK1 and RCLK185 complex to OsMKK4/5 in rice[21,22].There are 75 MAPKKK-like kinases in rice [23], however,their functions in disease immunity are largely unknown.
To elucidate how OsBSK1-2 regulates disease response in rice,we screened a yeast two-hybrid library and found two MAPKKKs,OsMAPKKK16 and OsMAPKKK18, that can interact with OsBSK1-2.We also found that OsBSK1-2 interacts with OsMAPKKK19, a homologue of OsMAPKKK16.All three MAPKKKs were induced after M.oryzae infection, and their overexpression in N.benthamiana leaves induced cell death.Moreover, we show that all three MAPKKKs function in regulation of response to blast in rice.They associate with OsMKK4/5, as well as OsBSK1-2 to mediate chitininduced MAPK activation.However, OsBSK1-2 was not involved in ETI.Our studies revealed key roles of the OsBSK1-2-OsMAPKK K16/18/19-OsMKK4/5 module in regulating disease response in rice.
2.Materials and methods
2.1.Plant materials and growing conditions
osmapkkk16, osmapkkk18 and osbsk1-2 mutants were constructed by the CRISPR/Cas9 method in wild-type Zhonghua 11(ZH11): the sgRNAs for osmapkkk16-1 and osmapkkk16-2 were 5′-GCGAGGTCGAGCGACTTGCG-3′and 5′-GGTGAACAAGTAGCAG GACC-3′, respectively; those for osmapkkk18-1 and osmapkkk18-2 were 5′-GGATTAACTGTGGACAACGG-3′and 5′-GCTCCAGTAGG GATTCGGTG-3′, respectively; and that for osbsk1-2 was 5′-GGGCG CCCAACCTCGTCTAC-3′.Plants overexpressing OsMAPKKK19 were generated by transforming a pCAMBIA1300-35S-OsMAPKKK19 plasmid into ZH11.All rice plants used for M.oryzae infection were grown under a 16-h light/8-h darkness photoperiod at 25–28 °C.
2.2.Yeast two-hybrid assays
Yeast two-hybrid assays were performed as described previously [24].Briefly, the full CDSs of the five OsMAPKKKs(8/11/16/18/19) were cloned into the pGADT7 vector to produce a prey plasmid, and OsBSK1-2 and OsMKK4/5 were cloned into the pGBKT7 vector to produce a bait plasmid.The bait and prey plasmids were co-transformed into yeast strain AH109 and grown on SD/-Leu-Trp (SD/-L-W) medium for 3 d.Positive clones were selected on SD/-Leu -Trp -His -Ade + X-α-gal (SD/-L-W-H-A + Xα-gal) medium.
2.3.Bimolecular fluorescence complementation (BiFC) assays
BiFC assays were performed as described previously [25].Briefly, OsMAPKKK16/18/19, OsMKK4/5 and OsBSK1-2 were fused with the N- or C-terminus of yellow fluorescent protein (YFP) by the Gateway method.Then, pairs of proteins fused with the Nterminus and C-terminus of YFP were co-expressed in N.benthamiana leaves by Agrobacterium-mediated transient transformation,and YFP signals were detected with a Zeiss 880 LSM confocal microscope.
2.4.Split-luciferase complementation imaging (LCI)
LCI assays were performed as described previously[26].Briefly,the two implicated genes were fused with the N-or C-terminus of luciferase and co-expressed in N.benthamiana leaves.Three d later,the leaves were sprayed with 1 mmol L-1luciferin,and fluorescent signals were detected by a low-light cooled CCD imaging apparatus.
2.5.Inoculation with the blast fungus
Inoculations using M.oryzae isolate Guy11 by different methods were performed as described previously[24].Leaves of two-weekold rice seedlings were sprayed with a spore suspension (5 × 105spores per mL), and the inoculated plants were placed in a dark humidity chamber for 24 h at 26 °C before transfer to a humid room with 14 h of light and 8 h of darkness at 26°C for two d,after which the diseased leaves were phenotyped.For punch inoculation,the leaves of one-month-old rice plants were lightly wounded with a punch and then 10 μL of spore suspension (5 × 105spores per mL)was added to the wound and sealed with plastic tape.The inoculated plants were placed in a dark humidity chamber for 24 h at 26 °C and then transferred to a humid room with 14 h of light and 8 h of darkness at 26 °C.Lesion length was measured 6 to 8 d after inoculation.
2.6.Real-time quantitative PCR (RT-qPCR)
Total RNA was isolated with TRIzol reagent (Invitrogen, Waltham, MA, USA), and total cDNA was synthesized with an All-in-One First-Strand cDNA Synthesis SuperMix for qPCR kit(TransGen Biotech, Beijing, China, AE341-02).Then, RT-qPCR was performed using a Green qPCR SuperMix kit(TransGen Biotech,Beijing,China,AQ601-01).The rice ubiquitin gene was used as the internal control.For fungal biomass assays, total DNA extracted from infected leaves was used for RT-qPCR,and relative fungal biomass was calculated as the amount of Pot2 gene in M.oryzae relative to ubiquitin in rice.Primers are listed in Table S1.
2.7.Cell death assay
Cell death assay was performed as described previously [22].The OsMAPKKK16/18/19 and GFP fusion proteins regulated by the 35S promoter were expressed in N.benthamiana leaves following Agrobacterium-mediated transient transformation, and photographs of the tissues were taken aft er 3 d.Tissues were stained with trypan blue as described previously [27].
2.8.Subcellular localization
To determine the subcellular localization of OsBSK1-2 and OsMAPKKK16/18/19, their full-length coding sequences were cloned separately into pCAMBIA1300-GFP vector, and the constructs were transformed into Agrobacterium strain GV3101 and injected into N.benthamiana leaves.GFP signals were assessed and photographed using a Zeiss 880 confocal microscope 36 h after injection.
2.9.Plant protein extraction and immunoblotting

Fig.1.OsBSK1-2 interacts with OsMAPKKK16 and OsMAPKKK18.(A,B)Yeast two-hybrid assay was used to determine the interactions of OsBSK1-2 with OsMAPKKK16 and OsMAPKKK18.Yeast cells containing the indicated plasmids were spotted onto SD/-L/-W and SD/-A/-H/-L/-W + X-α-gal mediums.Yeast cells harbouring OsBSK1-2-BD and OsMAPKKK16-AD or OsMAPKKK18-AD grew on SD/-A/-H/-L/-W + X-α-gal medium, but not the controls.(C, D) Split-luciferase complementation imaging assays used to determine interactios of OsBSK1-2 with OsMAPKKK16 and OsMAPKKK18 showed that constructs transiently expressed in N.benthamiana leaves were detected by fluorescent signals 3 d after injection.Fluorescent signal was detected when OsBSK1-CLUC was co-expressed with OsMAPKKK16-NLUC or OsMAPKKK18-NLUC,but not in controls.(E,F).Interactions of OsBSK1-2 with OsMAPKKK16 and OsMAPKKK18 were verified by bimolecular fluorescence complementation assay when OsBSK1, OsMAPKKK16 and OsMAPKKK18 were fused to the N-terminus(YN) or C-terminus(YC)of the YFP protein,respectively.Yellow fluorescence was detected when OsBSK1-YC was co-expressed with OsMAPKKK16-YN or OsMAPKKK18-YN, but not the controls.Scale bar, 20 μm.
Plant protein extraction was performed as described[28].Plant materials were quickly frozen in liquid nitrogen and ground to powder.The samples were incubated in plant protein extraction buffer containing 10% glycerol, 50 mmol L-1Tris-HCl (pH = 7.5),150 mmol L-1NaCl,2 mmol L-1EDTA,1 mmol L-1DTT,0.01%Triton X-100 and protease inhibitor cocktail for 30 min on ice.After centrifugation at 14,000 r min-1for 30 min at 4°C,the supernatant was transferred to a new tube for later use.Immunoblotting was performed as described with minor modifications [29].Proteins were separated by 10% SDS-polyacrylamide gel electrophoresis(PAGE) and transferred to a PVDF membrane.The membrane was incubated in TBST buffer containing 5% skim milk powder for 1 h at room temperature, followed by incubation overnight at 4 °C in TBST buffer containing 3%skim milk powder and the primary antibody.Finally,the membrane was placed in TBST buffer containing 3% skim milk powder and the secondary antibody for 1 h at room temperature.After washing with TBST buffer three times,the separated protein bands were detected with chemiluminescent HRP substrate by a chemiluminescence imaging system.
2.10.In vitro phosphorylation assays
In vitro phosphorylation assays were performed as described previously [30].Kinase and substrate proteins were expressed in E.coli and purified.Phosphorylation assays were performed at 30 °C for 30 min in a 30 μL reaction mixture containing 2.5 mmol L-1pH 7.5 Tris-HCl, 10 mmol L-1MgCl2, 10 mmol L-1CaCl2,10 mmol L-1MnCl2, 1 mmol L-1ATPγS, 1 mmol L-1dithiothreitol(DTT), 1 μg of kinase and 1 μg of substrate.Next, 2.5 mmol L-1pnitrobenzyl mesylate was added and the mixture was incubated at 30°C for 120 min.The reaction was terminated by addition of SDS sample buffer and incubated at 95°C for 10 min.The samples were analysed using immunoblotting with recombined antithiophosphate ester antibody.
2.11.MAPK activation assays
MAPK signaling activation assays were performed as described previously with minor modifications[22].Stems of 14-day-old rice seedings were cut into 3 mm segments and immersed in ddH2O overnight.After treating the stem tissue with 5 mmol L-1CaCl2for 30 min, 1 μmol L-1chitin (containing 0.02% Silwet L-77) was added and samples were collected after 0,5,15 and 30 min.MAPK activation was detected by immunoblotting with anti-p42/44 phosphorylation antibodies.
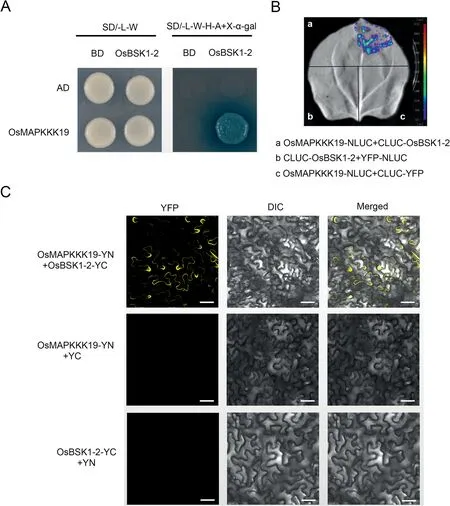
Fig.2.OsBSK1-2 interacts with OsMAPKKK19.(A)Yeast two-hybrid assay was used to determine the interaction of OsBSK1-2 and OsMAPKKK19.Yeast cells containing the indicated plasmids were spotted onto SD/-L/-W and SD/-A/-H/-L/-W+X-α-gal mediums.Yeast cells harbouring OsBSK1-2-BD and OsMAPKKK19-AD can grow on SD/-A/-H/-L/-W + X-α-gal mediums, but not the controls.(B) Split-luciferase complementation imaging assay was used to determine the interaction of OsBSK1-2 and OsMAPKKK19.Fluorescent signal was detected when CLUC-OsBSK1 was co-expressed with OsMAPKKK19-NLUC in N.benthamiana leaves, but not in the controls.(C) Bimolecular fluorescence complementation assay verified interaction of OsBSK1-2 and OsMAPKKK19.Yellow fluorescence was detected when OsBSK1-YC was co-expressed with OsMAPKKK19-YN in N.benthamiana leaves, but in not the controls.Scale bar, 20 μm.
3.Results
3.1.OsBSK1-2 was not involved in response to avirulent M.Oryzae strains
BSK1, a RLCK XII subfamily member, plays important roles in response to virulent Pseudomonas syringae pv tomato(Pto) strain DC3000 and avirulent Pto DC3000 strains that carry effector genes avrRpt2 or avrPphB in Arabidopsis [10].Silencing of OsBSK1-2, the homologue of BSK1,also decreased the level of resistance to M.oryzae in rice, [14].To determine whether OsBSK1-2 affects response to avirulent M.oryzae strains, we generated an osbsk1-2 knockout mutant in ZH11 by CRISPR/Cas9 (Fig.S1).Similarly, the osbsk1-2 knockout mutant showed decreased resistance to the virulent M.oryzae strain Guy11 compared to ZH11 (Fig.S1B,C).Avirulent M.oryzae strain JY14072 (which is virulent to pizh-1) was used to determine whether OsBSK1 was involved in a resistant response.It was reported that the Pizh-1 and Pizh-2 proteins combine to confer broad-spectrum disease resistance; Pizh-1 conferred resistance in ZH11,and Pizh-2 acted as a helper to promote Pizh-1 function [31].At 4 d post-inoculation with JY14072, disease lesions appeared in the pizh-1 mutant but not in the osbsk1-2 mutant and ZH11 (Fig.S1D).To validate this result, we used M.oryzae strain D527, which is avirulent to both ZH11 and the pizh-1 mutant,to inoculate the osbsk1-2 mutant.ZH11,and the pizh-1 and osbsk1-2 mutants were all resistant to strain D527, but control Co39 plants were not (Fig.S1E).These results indicated that OsBSK1-2 was not involved in resistance to avirulent M.oryzae strains.
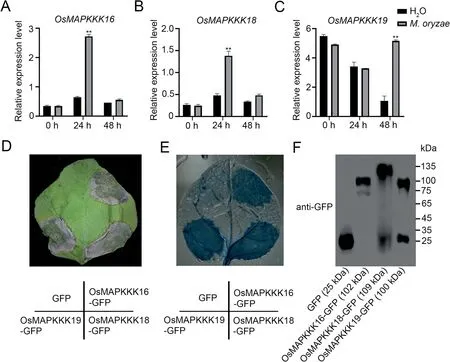
Fig.3.Expression of OsMAPKKK16, 18 and 19 after M.oryzae infection and cell death caused by their overexpression in N.benthamiana leaves.(A–C) Expression levels of OsMAPKKK16(A),OsMAPKKK18(B)and OsMAPKKK19(C)in 3-week-old wild-type rice plants inoculated with M.oryzae and sampled at 0,24 and 48 h.Values are means±SD(n=3)(**,P<0.01,Student’s t-tests).(D)Effects of GFP fusion proteins involving OsMAPKKK16,OsMAPKKK18 and OsMAPKKK19 expressed in N.benthamiana leaves;GFP was used as the negative control.Cell death was observed after three d.(E)Cell death confirmed by trypan blue staining and decoloring with chloral hydrate.(F)Immunoblots of GFP and the three MAPKKK-GFP fusion proteins.
3.2.OsMAPKKK16andOsMAPKKK18proteinsinteractedwithOsBSK1-2
The signaling pathway of OsBSK1-2 was unknown.Yeast twohybrid library screening was performed to identify the signaling pathway components of OsBSK1-2 (Table S2).From 40 identified candidates two MAPKKKs, OsMAPKKK16 and OsMAPKKK18, were selected for further studies because MAPKKKs were reported to transmit immune signals from RLCKs [11,21,22] and were identified more than once in the screen.Interaction was demonstrated when OsMAPKKK16 and OsMAPKKK18 were cloned into pGADT7 and co-transformed with OsBSK1-2-pGBKT7 into yeast (Fig.1A,B).A split-luciferase complementation imaging (LCI) assay was performed in N.benthamiana to further confirm these interactions.Luminescence signals were detected when OsBSK1-2 was cotransformed with each MAPKKK but not in the respective negative controls(Fig.1C,D).In addition,bimolecular fluorescence complementation (BiFC) assays indicated that OsBSK1-2 interacted with both MAPKKKs in the plasma membrane (Fig.1E, F).These results showed that that OsMAPKKK16 and OsMAPKKK18 interact with OsBSK1-2.
3.3.OsBSK1-2 interacted with OsMAPKKK19, a homologue of OsMAPKKK16
There are 75 MAPKKKs in rice, and phylogenetic analysis showed that OsMAPKKK8 and OsMAPKKK19 are homologues of OsMAPKK16, and OsMAPKKK11 is a homologue of OsMAPKKK18(Fig.S2).Therefore, we also determined whether OsBSK1-2 can interact with OsMAPKKK8,OsMAPKKK19 and OsMAPKKK11.Yeast two-hybrid assays showed that OsBSK1-2 interacted with OsMAPKKK19 (Fig.2A), but not with OsMAPKKK8 and OsMAPKKK11 (Fig.S3).An LCI assay performed in N.benthamiana verified interaction between OsBSK1-2 and OsMAPKKK19.Luminescent signals were detected when OsBSK1-2 and OsMAPKKK19 were co-expressed but were not present in the negative controls(Fig.2B).Interaction between OsBSK1-2 and OsMAPKKK19 was further confirmed by BiFC assay (Fig.2C).YFP signals were detected when OsBSK1-2 and OsMAPKKK19 were co-expressed in N.benthamiana, but were not detected when OsBSK1-2-YN and OsMAPKKK19-YC were co-expressed with YC or YN, respectively.The results showed that OsBSK1-2 interacts with OsMAPKKK19 in the plasma membrane.
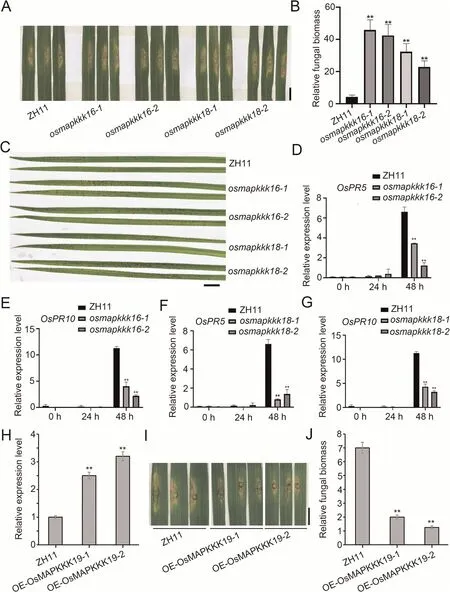
Fig.4.Knock out of OsMAPKKK16 and OsMAPKKK18 decreased resistance to M.oryzae;overexpression of OsMAPKKK19 increased resistance to M.oryzae.(A)Blast symptoms in ZH11, and osmapkkk16 and osmapkkk18 knockout mutants 7 d after inoculation with M.oryzae isolate Guy11 by the punch method.Scale bar, 1 cm.(B) Relative fungal growth in lesions from ZH11 and osmapkkk16 and osmapkkk18 mutants by qRT-PCR at the DNA level with M.oryzae Pot2 gene compared to rice ubiquitin.(**, P < 0.01,Student’s t-test).(C)Twenty-day-old wild type(ZH11),and osmapkkk16 and osmapkkk18 mutant plants were inoculated with M.oryzae isolate Guy11 by the spray method.The disease lesions were observed 3 d after inoculation.More lesions were observed in osmapkkk16 and osmapkkk18 mutant plants than in ZH11.Scale bar, 1 cm.(D–G)Expression levels of PR genes OsPR5 and OsPR10 in the ZH11 and osmapkkk16 and osmapkkk18 mutants at indicated times after inoculation with M.oryzae.(**, P < 0.01,Student’s t-tests).(H) OsMPKKK19 expression in one-month-old OE-OsMPKKK19 plants with ZH11 as control.(I) Lesions of one-month-old ZH11 and OE-OsMPKKK19 mutants 7 d after punch-inoculation with the M.oryzae isolate Guy11.Scale bar, 1 cm.(J) Relative fungal biomass in disease lesions in ZH11 and OE-OsMPKKK19 7 d postinoculation with M.oryzae Pot2 gene compared to rice ubiquitin.(**, P < 0.01, Student’s t-test).
3.4.Expression of OsMAPKKK16,18 and 19 was induced by M.Oryzae infection, and their respective overexpression induced cell death in N.Benthamiana leaves
OsBSK1-2 was reported to regulate blast response in rice, and given that OsMAPKKK16, 18 and 19 can interact with OsBSK1-2,we speculated that OsMAPKKK16/18/19 may also be involved in immune responses.Studies of the three MAPKKKs following inoculation of rice seedlings with M.oryzae indicated that expression of all three MAPKKKs was induced by M.oryzae infection(Fig.3A–C).OsMAPKKK16 and OsMAPKKK18 were significantly upregulated at 24 h whereas OsMAPKKK19 was upregulated at 48 h post-inoculation infection,but not after treatment with water.Since some immunity-related proteins, including OsMAPKKK18,were known to induce cell death in N.benthamiana leaves[11,21], we expressed OsMAPKKK16 and OsMAPKKK19, driven by the 35S promoter, in N.benthamiana leaves using OsMAPKKK18 as the positive control and GFP as the negative control.All four proteins were expressed in N.benthamiana leaves (Fig.3F), and like OsMAPKKK18, both OsMAPKKK16 and OsMAPKKK19 but not the negative control induced cell death (Fig.3D,E).These results indicated that OsMAPKKK16, 18 and 19 were involved in immune response.
Subcellular localization in N.benthamiana leaves showed that OsBSK1-2 and the three OsMAPKKKs were mainly located on the cell membrane whereas GFP alone was located throughout the entire cell (Fig.S4).
3.5.Knockout of OsMAPKKK16 and OsMAPKKK18 compromised resistance to M.Oryzae
Pairs of osmapkkk16 and osmapkkk18 mutants generated by CRISPR/Cas9 in ZH11 (Fig.S5) were used to further investigate the roles of the respective wild type alleles in immunity.When the four mutants and ZH11 grown for 4–5 weeks were inoculated with M.oryzae Guy11 by the punch method,the lesion areas in the mutants were larger than those in the wild type (Fig.4A).Fungal biomass was higher in leaves of the mutants compared to the wild type (Fig.4B).When the five lines were inoculated by the spray method, the mutants produced more disease lesions than ZH11(Fig.4C).Furthermore, the transcript levels of pathogenesisrelated genes OsPR5 and OsPR10 in infected leaves at 48 h postinoculation were lower in the mutants than in the wild type(Fig.4D–G).The results above suggested that OsMAPKKK16 and OsMAPKKK18 were involved in response to M.oryzae.Some agronomic traits were also changed in the mutants;for example,grain length and thousand grain weight were increased in the osmapkkk16 mutants and grain width and thousand grain weight were decreased in the osmapkkk18 mutants compared to ZH11(Fig.S6), indicating multiple roles of these genes in disease response and plant development.
3.6.Overexpression of OsMAPKKK19 increases resistance to M.Oryzae
Selected lines OE-OsMAPKKK19-1 and OE-OsMAPKKK19-2 in which expression of OsMAPKKK19 was 2.5 and 3.1 times, respectively, higher than that ZH11 (Fig.4H) inoculated with M.oryzae by the punch method had reduced lesion lengths (Fig.4I).Additionally, fungal growth in the lesions of the overexpression lines was less than in ZH11 (Fig.4J).These results suggested that overexpression of OsMAPKKK19 increased resistance to M.oryzae.Moreover,overexpression of OsMAPKKK19 had no significant effect on plant growth and development (Fig.S7).
3.7.OsMAPKKK16 and OsMAPKKK18 interacted with, and phosphorylated, OsMKK4 and OsMKK5
MKK4 and MKK5 are MAPK cascade members that transmit immune signals from MAPKKK to MAPK proteins [32,33].In rice,OsMKK4 and OsMKK5 deliver chitin-triggered immune signals from OsMAPKKK18 and OsMAPKKKε to OsMPK3/6 [21,22].We hypothesised that OsMAPKKK16, OsMAPKKK18 and OsMAPKKK19 might transmit immune signals downstream from OsBSK1-2 by a OsMKK4/OsMKK5-OsMPK3/OsMPK6 cascade.First, we showed that both OsMAPKKK16 and OsMAPKKK19 interacted with OsMKK4 and OsMKK5 (Fig.5).Since OsMAPKKK18 was reported to interact with and phosphorylate OsMKK4[21]we predicted that OsMAPKKK16 and OsMAPKKK19 would also phosphorylate OsMKK4 and OsMKK5.An in vitro phosphorylation assay to verify this hypothesis confirmed that both OsMAPKKK16 and OsMAPKKK19 phosphorylated OsMKK4 and OsMKK5(Fig.6).Since RLCKs generally transmit immune signals by phosphorylation of substrates to regulate their function we deduced that OsBSK1-2 may phosphorylate all three OsMAPKKKs.So the autophosphorylation activity of OsBSK1-2 was detected.However, we could not detect the autophosphorylation activity of OsBSK1-2 (Fig.S8).
3.8.The OsMAPKKK16/18/19-OsMKK4/5-OsMPK3/6 cascade causes OsBSK1-2-mediated immune signal transduction
Since OsMAPKKK18 activates MAPK [21] we speculated that OsMAPKKK16,OsMAPKKK19 and OsBSK1-2 could also be involved in MAPK activation.For verification we examined chitin-activated MAPK signals in osmapkkk16 and osbsk1-2 mutants and used ZH11 as the control.As shown in Fig.7A and B activation of OsMPK3 and OsMPK6 was reduced in the mutants suggesting that the OsMAPKKK16/18/19-OsMKK4/5-OsMPK3/6 module regulated OsBSK1-2-mediated immune signal transduction.
4.Discussion
We identified three MAPKKK proteins (OsMAPKKK16,OsMAPKKK18 and OsMAPKKK19) that interacted with OsBSK1-2(Figs.1,2).Expression of all three was induced by M.oryzae infection,and their overexpression in N.benthamiana leaves caused cell death(Fig.3),indicating roles in regulation of immunity.Knockout of OsMAPKKK16 and OsMAPKKK18 reduced the level of resistance,and overexpression of OsMAPKKK19 increased resistance as measured by lesion size and fungal growth (Fig.4), suggesting that OsMAPKKK16, OsMAPKKK18 and OsMAPKKK19 are involved in blast response.Like OsMAPKKK18, OsMAPKKK16 and OsMAPKKK19 interacted with and phosphorylated OsMKK4 and OsMKK5 (Figs.5, 6), with activation of the MAPK signaling pathway being lower in osmapkkk16 and osbsk1-2 mutants(Fig.7),thus indicating that OsBSK1-2 functions by activating MAPK signaling through multiple MAPK signaling pathways.
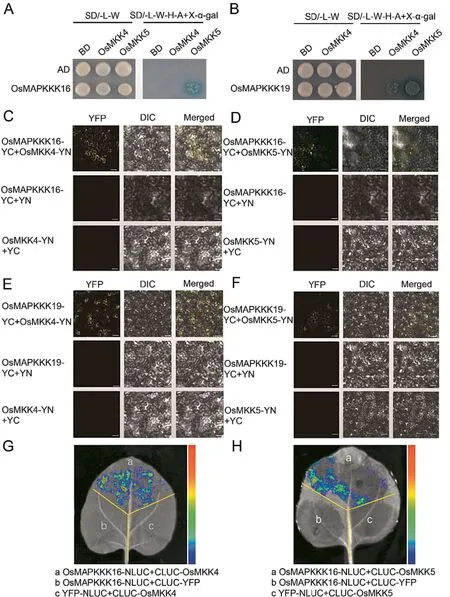
Fig.5.Interactions of OsMAPKKK16 and OsMAPKKK19 with OsMKK4 and OsMKK5.(A, B) Y2H assays were used to determine interactions of OsMAPKKK16 (A) and OsMAPKKK19(B)with OsMKK4 and OsMKK5.Yeast cells harbouring OsMKK5-BD and OsMAPKKK16-AD,as well as OsMKK4-BD or OsMKK5-BD and OsMAPKKK19-AD grew on both SD/-L/-W and SD/-L-W-H-A+X-α-gal media,but not others.(C,D)BiFC assays showed that OsMAPKKK16 interacted with OsMKK4 and OsMKK5.Yellow fluorescence was detected when OsMAPKKK16-YC and OsMKK4-YN or OsMKK5-YN were co-expressed in N.benthamiana leaves,but not the controls.Scale bar,20 μm.(E,F)BiFC assays showed that OsMAPKKK19 interacted with OsMKK4 and OsMKK5.Yellow fluorescence was detected when OsMAPKKK19-YC and OsMKK4-YN or OsMKK5-YN were coexpressed in N.benthamiana leaves,but not in the controls.Scale bar,20 μm.(G,H)LCI assays showing interactions of OsMAPKKK16 with OsMKK4 and OsMKK5.Fluorescent signals were detected when CLUC-OsMKK4 or CLUC-OsMKK5 was co-expressed with OsMAPKKK16-NLUC in N.benthamiana leaves, but not in the negative controls.
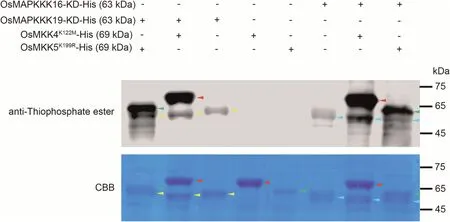
Fig.6.In vitro phosphorylation assay of OsMAPKKK16 and OsMAPKKK19 on OsMKK4 and OsMKK5.The kinase domains of OsMAPKKK16 and OsMAPKKK19(OsMAPKKK16-KD and OsMAPKKK19-KD),as well as inactive kinase versions OsMKK4K122M and OsMKK5K99R,were cloned and expressed in E.coli strain BL21.In vitro phosphorylation assay was performed with indicated recombinant proteins and immunoblotted with anti-thiophosphate ester antibody.Input recombinant proteins were detected by Coomassie brilliant blue (CBB) staining.Yellow arrows, OsMAPKKK19-KD; blue arrows, OsMAPKKK19-KD; red arrows, OsMKK4K122M; green arrows, OsMKK5K99R.
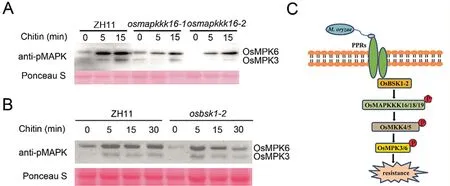
Fig.7.MAPK activation assays and proposed working model.(A) MAPK activation assays in ZH11 and osmapkkk16 mutant.Stems of ten-day-old ZH11 and osmapkkk16 seedlings were treated with 1 μmol L-1 chitin.MAPK activation was detected at indicated time points using immunoblotting with anti-p42/44 phosphorylation antibodies.(B) MAPK activation assays in ZH11 and osbsk1-2 mutant.Stems of ten-day-old ZH11 and osbsk1-2 seedlings were treated with 1 μmol L-1 chitin.MAPK activation was detected as described above.(C) Functional model of the OsBSK1-2-OsMAPKKK16/18/19-OsMKK4/5 module in rice.Following M.oryzae infection PRRs transmit immune signals to OsBSK1-2, and OsBSK1-2 affects MAPK activation via OsMAPKKK16/18/19 mediated OsMKK4/5 phosphorylation.
BSK1 belongs to the RLCK XII subfamily and has been reported to play important roles in regulation of plant immunity[10,12,14].In Arabidopsis, where the functional mechanism of BSK1 has been well demonstrated BSK1 receives immune signals from FLS2 and RLK902 [10,12] and transmits immune signals to MAPKKK5 and MPK15 [11,13].BSK1 balances growth and immunity by dynamic spatial reorganization [34].OsBSK1-2, an orthologue of BSK1, is involved in regulation of blast resistance in rice [14] but its functional mechanism is unknown.We identified MAPKKK proteins OsMAPKKK16, OsMAPKKK18 and OsMAPKKK19, that interacted with OsBSK1-2 (Figs.1, 2).Bioinformatics analysis revealed that there are 75 MAPKKKs in rice[23]and some were reported to function in regulation of immunity.For instance, OsMAPKKKε was involved in OsCERK1- and RLCK185-mediated chitin signal transduction and knockdown of OsMAPKKKε compromised resistance to M.oryzae [22].OsEDR1, a MAPKKK family protein, negatively regulated bacterial resistance by inhibiting OsMPKK10.2 activity,which is also negatively regulated by OsMPK6 [35].Rice Raf-like MAPKKK OsILA1 is involved in bacterial blight resistance by suppressing the OsMAPKK4–OsMAPK6 cascade [36].The present results showed that three OsBSK1-2-interacting proteins,OsMAPKKK16, OsMAPKKK18 and OsMAPKKK19, are involved in regulation of blast response (Fig.4).OsMAPKKK18 is a homologue of AtMAPKKK5, and OsMAPKKK16 and OsMAPKKK19 are orthologues of AtMAPKKK3 (Fig.S2).In Arabidopsis, BSK1 regulates immunity via AtMAPKKK5 [11].In apple, MdMAPKKK1, a homologue of AtMAPKKK5, interacts with MdBSK1-1 and MdBSK1-2 and positively regulates resistance to Botryosphaeria dothidea[37].Finally, AtMAPKKK5 and AtMAPKKK3 are key proteins that bridge RLCK VII members to the MAPK signaling pathway [38].Our results reveal that the signaling pathway of RLCK VII members in immunity regulation in rice is relatively conserved.However,there are more MAPKKK homologues of AtMAPKKK5 and AtMAPKKK3 in rice (Fig.S2), indicating functional differentiation between rice and other plants.Whether OsMAPKKK8 and OsMAPKKK11, along with OsMAPKKK9 and OsMAPKK10, are also involved in regulation of immunity and whether OsMAPKKK16,OsMAPKKK18 and OsMAPKKK19 are involved in immune signaling pathways mediated by other RLCKVII members requires further investigation.
BSK1 phosphorylates MAPKKK5 at Ser-289, and phosphorylation of the Ser-289 residue is critical for MAPKKK5-mediated resistance to both bacterial and fungal pathogens in Arabidopsis [11].Since we deduced that OsBSK1-2 phosphorylates all three OsMAPKKKs we first tested for autophosphorylation activity of OsBSK1-2 and found none (Fig.S7).Some kinases phosphorylate downstream substrates but require pre-activation by upstream kinases [35,39,40].It seems that activation of OsBSK1-2 may depend on upstream receptor or receptor-like kinases (RKs or RLKs); a feature worth clarifying in the future.
The MAPK signaling pathway was reported to play important roles in regulation of plant immunity, and the MKK4/MKK5-MPK3/MPK6 cascade is a conserved module that links MAPKKK proteins and downstream defence-related proteins [20,38].A previous report showed that OsMAPKKK18 interacts with and phosphorylates OsMKK4 to affect MAPK activation [21].In this study,we found that both OsMAPKKK16 and OsAMPKKK19 interact with and phosphorylate OsMKK4 and OsMKK5 (Figs.5, 6), indicating that OsBSK1-2 functions through the OsMAPKKK16/18/19-OsMK K4/5-OsMPK3/6 cascade.Consistent with this finding, activation of OsMPK3 and OsMPK6 was lower in the osbsk1-2 mutant than the wild type (Fig.7B).Interestingly, MAPK activation was not altered in the Arabidopsis bsk1 mutant [10], which displayed enhanced susceptiblity to virulent Pto strain DC3000 and avirulent Pto DC3000 strains that carry the effector genes avrRpt2 or avrPphB.However,in rice,the osbsk1-2 mutant exhibited enhanced susceptiblity to virulent strains of M.oryzae but the same level of resistance to an avirulent strain compared to ZH11 (Fig.S1), indicating functional differentiation of BSK1 in rice and Arabidopsis.Moreover, OsMAPKKK16 and OsMAPKKK19 interacted with OsMKK4 and OsMKK5, indicating that OsBSK1-2 may function in the same complex.Further studies are required to investigate interaction of OsBSK1-2 with OsMKK4 and OsMKK5.
We showed that OsMAPKKK16 and OsAMPKKK19 can phosphorylate OsMKK4 and OsMKK5 in vitro (Fig.6).OsMAPKKK18 was previously reported to phosphorylate OsMKK4 [21].Furthermore,three OsMAPKKKs interacted with OsBSK1-2, a RLCK protein.Whether the OsMAPKKKs phosphorylation of OsMKK4/5 depends on OsBSK1-2 in vivo remains unknown.The differences in phosphorylation of OsMKK4/5 by OsMAPKKKs in the wild-type and osbsk1-2 mutant will be investigated in future work.
In addition to regulating immunity, OsMAPKKK16 and OsMAPKKK18 affect grain size and thousand-grain weight in rice(Fig.S6).The OsMKKK10-OsMKK4-OsMAPK6 signaling pathway also positively regulates grain size and weight in rice[39].Interestingly,overexpression of OsMAPKKK19 induced cell death in N.benthamiana leaves but did not affect the growth and development of rice.One possibility for this result was that the expression level of OsMAPKKK19 in overexpressing plants was less than threefold compared to the wild type(Fig.4H),and not as high as when transiently expressed in N.benthamiana.
In conclusion, our results demonstrated that an OsMAPKKK16/18/19-OsMKK4/5-OsMPK3/6 cascade was involved in OsBSK1-2-mediated immune signal transduction.As shown in Fig.7C, when rice was exposed to pathogen infection, several MAPK signaling pathways were activated through OsBSK1-2 and several MAPKKKs, activated downstream defence responses thereby reducing pathogen growth.
Gene accession numbers
Sequence data in this article can be found in the Rice Genome Annotation Project database (https://rice.uga.edu/) under the following accession numbers: OsBSK1-2 (LOC_Os10g39670),OsMAPKKK16 (LOC_Os04g35700), OsMAPKKK18 (LOC_Os03g55560),OsMAPKKK19 (LOC_Os02g35010), OsMAPKKK8 (LOC_Os11g10100),OsMAPKKK11 (LOC_Os07g02780), OsMKK4 (LOC_Os02g54600),OsMKK5 (LOC_Os06g09180), OsPR5 (LOC_Os03g46070), OsPR10(LOC_Os12g36880), and ubiquitin (LOC_Os03g13170).The sequence of the Pot2 gene can be found in the National Center for Biotechnology Information under accession number Z33638.
CRediT authorship contribution statement
Shengping LiandDingzhong Tang:conceived and designed the research.Xinquan Xiang, Shengping Li, Zhijuan Diao, Na Xia, Ling LuandJing Zhang:conducted most of the experiments.Shengping Li, Xinquan XiangandZhiwei Chen:data analysis.Shengping LiandDingzhong Tang:wrote the manuscript.All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2022YFF1001500) and Key Program of Technology and Science in Fujian province(2020NZ08016).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.11.009.
杂志排行
The Crop Journal的其它文章
- Flowering-time regulation by the circadian clock: From Arabidopsis to crops
- Global characterization of OsPIP aquaporins reveals that the H2O2 transporter OsPIP2;6 increases resistance to rice blast
- Drought-triggered repression of miR166 promotes drought tolerance in soybean
- Natural variation of an autophagy-family gene among rice subspecies affects grain size and weight
- Rice gene OsUGT75A regulates seedling emergence under deep-sowing conditions
- A telomere-to-telomere genome assembly of Zhonghuang 13,a widely-grown soybean variety from the original center of Glycine max
