MLST analysis of genetic diversity of Bacillus coagulans strains to evaluate effects on constipation model
2022-06-20JiangCaoWenyinLiuRuolanLiliuJianxinZhaoHaoZhangWeiChenQixiaoZhai
Jiang Cao, Wenyin Liu, Ruolan Liliu, Jianxin Zhao,Hao Zhang,c,d, Wei Chen,c, Qixiao Zhai,*
a State Key Laboratory of Food Science and Technology, Jiangnan University, Wuxi 214122, China
b School of Food Science and Technology, Jiangnan University, Wuxi 214122, China
c National Engineering Research Center for Functional Food, Jiangnan University, Wuxi 214122, China
d Wuxi Translational Medicine Research Center and Jiangsu Translational Medicine Research Institute Wuxi Branch, Wuxi 214122, China
Keywords:
Bacillus coagulans
Multilocus sequence typing
Housekeeping gene
Constipation
Strain-specific
A B S T R A C T
Bacillus coagulans can help ameliorate or prevent gastrointestinal diseases, but the genetic relationships among B. coagulans isolates are not well studied. Multilocus sequence typing analysis was conducted on 57 isolates of B. coagulans from 22 provinces or autonomous regions in China. B. coagulans isolates were highly diverse and a total of 33 (sequence typings) STs were found. These isolates had a weak clonal population structure and strong indications of intraspecies recombination. The evolution direction of B. coagulans was not correlated with geography or isolation source. Fifteen strains were selected for further analysis based on proximity relationships from the phylogenetic tree. Five isolates (B. coagulans-1, B. coagulans-10,B. coagulans-39, B. coagulans-70 and B. coagulans-71) with good spore-forming ability relative to the rest of the isolates were evaluated for constipation relief. B. coagulans-39 significantly relieved constipation symptoms in mice by regulating intestinal flora, increasing the production of short-chain fatty acids and restoring the level of gastrointestinal regulatory peptides. Comparative genomic analysis showed the beneficial effects of B. coagulans-39 might be associated with specific functional genes that are involved in the utilization of various carbohydrates as primary substrates and short-chain fatty acid production.
1. Introduction
Bacillus coagulanswas originally isolated by Hammer (1915)from canned milk, as a kind of lactic acid-producing bacterium associated with canned spoilage. This species was first identified asLactobacillus sporogenesbecause it had characteristics of spore formation and lactic acid production [1]. Genetic studies have confirmed thatB. coagulansshould be classified within the genusBacillus[2]. Isolates ofB. coagulansare distributed in a variety of habitats, including fermented foods, fermented vegetables,rhizosphere soil, canned food, and sewage [3].B. coagulansproduces a lot of metabolites, including short-chain fatty acids (SCFAs),L-lactic acid, and bacteriocins [4-7]. Similar to other reportedBacillusspecies,B. coagulanscan be used in the functional food fermentation [8]. This species is generally recognized as safe (GRAS) by the United States Food and Drug Administration (FDA) and European Union Food Safety Authority (EFSA).
Multilocus sequence typing (MLST), a molecular typing method based on housekeeping genes, is commonly used for the identification and characterization of bacterial species [9]. In contrast to otherBacillusspecies such asB. subtilis,B. cereusandB. thuringiensis, no MLST profiles ofB. coagulanswere found in the databases of PubMLST (https://pubmlst.org/databases). Based on the NCBI genome database (https://www.ncbi.nlm.nih.gov/),only 38B. coagulansstrain genomes have been sequenced, far fewer than those of otherBacillusspecies such asB. subtilis(377) andB. cereus(1122). Until now, published studies have identifiedB. coagulansas a heterogeneous species with high intra-species genomic diversity [10,11]. However, the dearth of genomic information onB. coagulanshas greatly limited the understanding of this species.
B. coagulanshas been observed to benefit human health in many studies [12,13].B. coagulanshas probiotic abilities, such as improving nutrient absorption [14], modulating gut microbiota [15],and immune responses [16]. Several supplementation trials indicate that ingestion ofB. coagulanshelps relieve constipation symptoms in mice and humans [17-20]. Constipation is a common gastrointestinal disease and a predisposing factor for many conditions with high death rates. Traditional therapeutic methods, such as behavioral intervention therapy and irritant drugs are generally used to prevent defecation, but these treatments have side effects [21]. Therefore, it is valuable to attempt treatment of constipation withB. coagulansas an intestinal micro-ecological therapy method. However, up to now, howB. coagulansrelieves constipation remains obscure. To the best of our knowledge, this study is the first to consider whetherB. coagulansexerts inter-strain differences in alleviating constipation.
A novel MLST scheme was developed to characterize the diversity, genetic population structure and evolutionary origins ofB. coagulansisolates. TheB. coagulansisolates selected from the MLST scheme were evaluated against constipation in mouse model.The genomic and functional characteristics of differentB. coagulansisolates were analyzed to assess their beneficial effects from genetic background.
2. Materials and methods
2.1 Chemicals and reagents
Bacteria genomic DNA extraction kits were purchased from Beijing Tiangen Biotechnology, China. Trypsin was purchased from Sangon Biotechnology, China. Bile salt was purchased from Bioleaf,China. Loperamide hydrochloride capsules were purchased from Xian Janssen Pharmaceutical Ltd., China. Activated carbon powder and pepsin were purchased from Sangon Biotechnology, China. All enzyme immunosorbent assays (ELISA) kits used in this study were purchased from Nanjing SenBeiJia Biological Technology, China.Arabic gum powder, acetic acid, propionic acid,n-butyric acid and isobutyric acid were purchased from Sinopharm Group Chemical Reagent Company, China. Fast DNA spin kits for feces genome extraction were purchased from MP company, USA. QIAquick Gel Extraction Kits were purchased from QIAGEN, Germany.Quant-iT PicoGreen dsDNA Assay Kits were purchased from Life Technologies Company, USA. Experimental ink was prepared according to the literature method [21]. de Man, Rogosa and Sharpe medium, nutrient yeast extract salt medium and nutrient-limited medium were prepared as previously described [22,23].
2.2 Preparation of spores and vegetative cells
B. coagulansstrains used in this study (Table S1) were preserved in the in-house Culture Collections of Food Microbiology, Jiangnan University (Wuxi, China). To prepare active cultures for the experiments, all strains were cultured under aerobic conditions for 24 h at 37 °C in MRS and consecutively reactivated three times before use. To prepare spores ofB. coagulansfor all experiments,the activated strains were inoculated into nutrient yeast extract salt medium and nutrient-limited medium with a 2% (V/V) inoculation amount. The strains were then cultured in flasks at 37 °C and 200 r/min for 48 h to collect the spore suspension.
2.3 LST analysis of B. coagulans
2.3.1 DNA extraction
The genome ofB. coagulanswas extracted using the bacterial genomic DNA extraction kit (Beijing Tiangen Biotechnology Co.,Ltd., China).
2.3.2 Selection of housekeeping genes for MLST
Seven housekeeping genes (gmk,tpi,ilvd,pur,pta,glpf,ldh)were selected based on the gene sequences ofB. coagulansATCC 7050. The housekeeping genes were selected from different regions in theB. coagulansATCC 7050 genome sequence (Table S2):tpi(triosephosphate isomerase),gmk(guanylate kinase, putative),pur(phosphoribosylaminoimidazole carboxamide),pta(phosphate acetyltransferase),ilvd(dihydroxy-acid dehydratase),glpf(glycerol uptake facilitator protein),ldh(lactate dehydrogenase).
2.3.3 PCR amplification and nucleotide sequencing
The primers of the housekeeping genes were designed with Primer 6.0 software (Table S2). Housekeeping genes ranged from 444-704 bp (Fig. S1). The polymerase chain reaction (PCR) mixture(25 μL) included 1 μL template, 12.5 μL 2 ×TaqMaster Mix (Beijing Tiangen Biotechnology Co., Ltd, China), 11 μL ddH2O, 0.25 μL Primer-F, and 0.25 μL Primer-R. The PCR procedure included the following conditions: 94 °C for 5 min; 35 cycles of amplification for 95 °C for 1 min; annealing for 45 s; 72 °C for 1 min; and extending at 72 °C for 10 min. The annealing temperatures of different genes were con firmed by continuous trials (Table S2). Sequences of each isolate were verified and organized as previously described [24].
2.3.4 Descriptive analysis of MLST sequence data
After theB. coagulanshousekeeping genes were sequenced and imported into BioNumerics 7.2 software, the allele number was formed according to the order of “tpi-gmk-pur-pta-ilvd-glpf-ldh”for each housekeeping gene locus (the order of housekeeping genes was automatically generated by BioNumerics 7.2 software). Seven allele numbers were combined into a sequence pattern corresponding to its strain. The polymorphic loci, G + C% content, dN/dSvalue,linkage disequilibrium analysis, and recombination analysis were performed as previously described [25]Bag. The assignment of ST to clone complex (CC) was achieved using Global optimal eBURST(goeBURST) with triple locus variant (TLV) limitation. A minimumspanning tree analysis was constructed using the Prim’s algorithm in BioNumerics 7.2 software. The phylogenetic tree ofB. coagulansisolates was constructed using the UPGMA algorithm in MEGA X software. analysis of variance (ANOVA) and correlation analysis betweenB. coagulansSTs and geography or isolation source were determined using the SPSS software.
2.4 Evaluation of physiological characteristics of B. coagulans in vitro
The sporulation abilityB. coagulanswas tested as described previously [26]. The total viable counts of the strains before and after heat treatment were determined by the count plate method.The sporulation rate of theB. coagulansstrains was calculated according to the counting results. The tolerance ofB. coagulansstrains to a simulated gastrointestinal environment was studied as previously described [27]. The survival rate ofB. coagulansspores and vegetative cells in the simulated gastrointestinal environment was measured based on the counting results. The germination ability of eachB. coagulansstrain in the simulated gastrointestinal environment was determined as described previously [28].
2.5 Animal experiment
2.5.1 Experimental design
All protocols for animal experiments in this study were approved by the Ethics Committee of Jiangnan University, China (JN.No20180430b1000801). Experiments were performed in accordance with European Community guidelines (Directive 2010/63/EU).BALB/c mice (7 weeks old, male, SLAC Laboratory Animal Co.,Ltd, Shanghai, China) were raised in the SPF level experimental animal barrier system of the experimental animal center of Jiangnan University. The environment was maintained at a temperature of(23 ± 2) °C and relative humidity of (50 ± 10)%. The mice were acclimatized for one week before the experiment.
The effects ofB. coagulansstrains on loperamide-induced constipation in mice were evaluated. The procedure for the animal experiment is shown in Table 1. The dose of loperamide was selected based on a previous study [20]. The animal trial lasted for 14 days.At the end of the experiment, the mice were euthanized for tissue and blood collection.
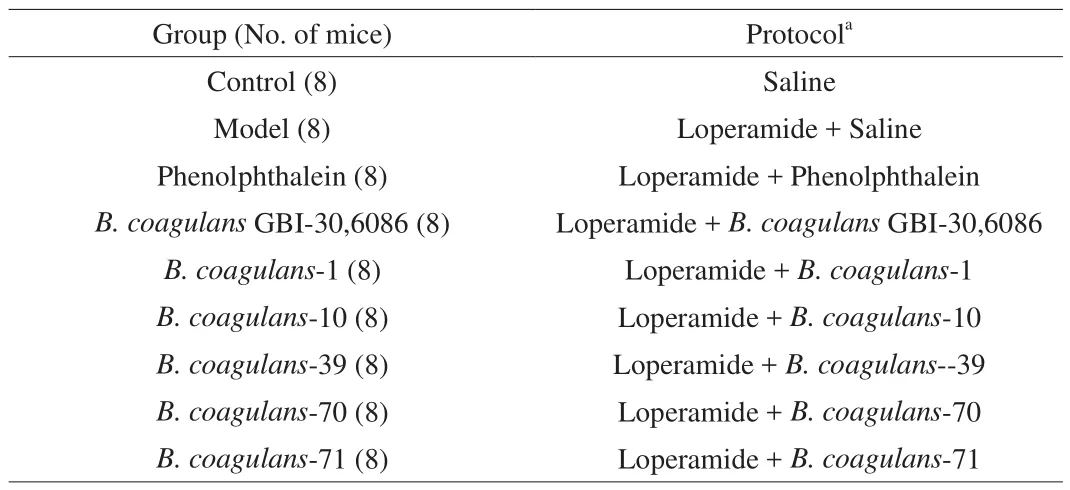
Table 1Animal experimental protocol.
2.5.2 Measurement of mice defecation status
The mice were immediately transferred into a single cage for 3 h after the end of the gavage on days 1, 7 and 14 to collect feces. The number of fecal particles and the wet and dry weights of feces were recorded. After freeze-drying, the water content was calculated as the difference between the wet and dry weights of the feces.
2.5.3 Determination of gastrointestinal transit ability
The time to the first black stool defecation and small intestinal propulsion rates of mice were determined based on a method adapted from Wang et al. [21].
2.5.4 Determination of short chain fatty acid contents in feces
The concentrations of SCFAs (acetate acid, propionate acid,butyrate acid and total acid) in the freeze-dried feces were analyzed by gas chromatography coupled mass spectrometry as previously described [29].
2.5.5 Genome sequencing analysis of community structures
The bacterial genomic DNA of fecal samples was extracted with a Fast DNA Spin Kit for Feces Kit. The amplification of the 16S rRNA gene sequence (V3–V4 regions) was performed as described previously [30].
2.5.6 Determination of serum gastrointestinal regulatory peptide
Serum parameters were determined by an ELISA instrument as previously described [21].
2.5.7 Comparative genomic analysis of B. coagulans
The genome sequencing and annotation of fiveB. coagulansisolates were performed as in previous studies [31-34]. Orthologous genes were generated using OrthoMCL1.4 with default parameters [35].Genome similarities were assessed by calculating the average nucleotide identity using BLAST (ANIb) in JSpecies [36].
2.5.8 The growth curves of B. coagulans
B. coagulansgrowth curves were measured in carbon limited medium containing glucose (5 mg/mL) and galatooligosaccharide(GOS) (5 mg/mL), as previously described [37].
2.6 Statistical analysis
Date are presented as mean ± standard (SD). The differences between the mean values for the groups were analyzed using a one-way ANOVA with Duncan’s multiple range test.P< 0.05 is considered to indicate statistical significance. All statistical analyses were performed and visualized with GraphPad Prism (GraphPad Software Inc., San Diego, CA, USA) and R scripts.
3. Results
3.1 MLST analysis of B. coagulans isolates
3.1.1 Sequence diversity in B. coagulans
General information about the housekeeping genes (tpi,gmk,pur,pta,ilvd,glpf,ldh) and primers used for MLST is given in Table S2.The analysis of genetic characteristics, including the size of gene,Pivalue, guanine-cytosine (GC) content and dN/dSvalue, indicated that high nucleotide diversity of individual housekeeping genes existed in theB. coagulansstrains (Table 2).
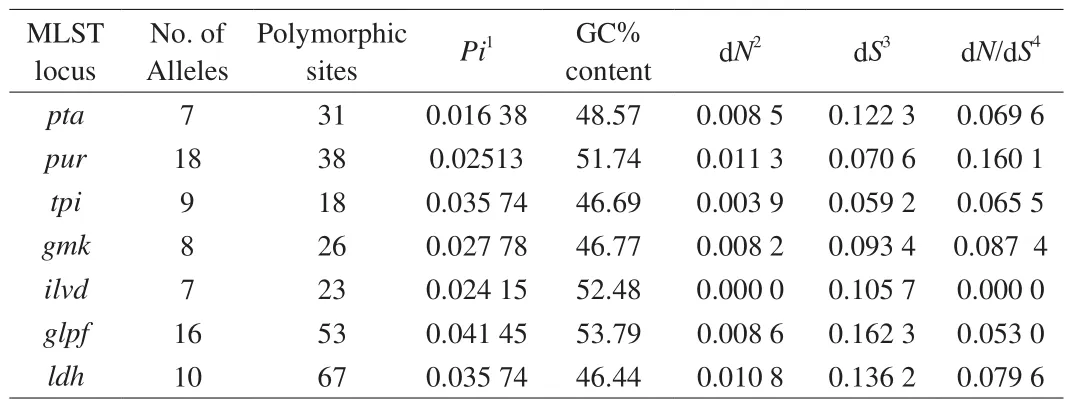
Table 2Nucleotide diversity of housekeeping genes in B. coagulans.
3.1.2 Recombination and linkage disequilibrium analysis of B. coagulans
TheIA(the index of association) andIAS(the standardized index of association) values were calculated as 0.789 2 and 0.131 5,respectively. Split decomposition analysis indicated that intergenic recombination occurred in the evolutionary history of all selected genes (Fig. 1A). In addition, the 33 STs ofB. coagulansshowed a complicated network structure with parallelogram-shaped structures(Fig. 1B).
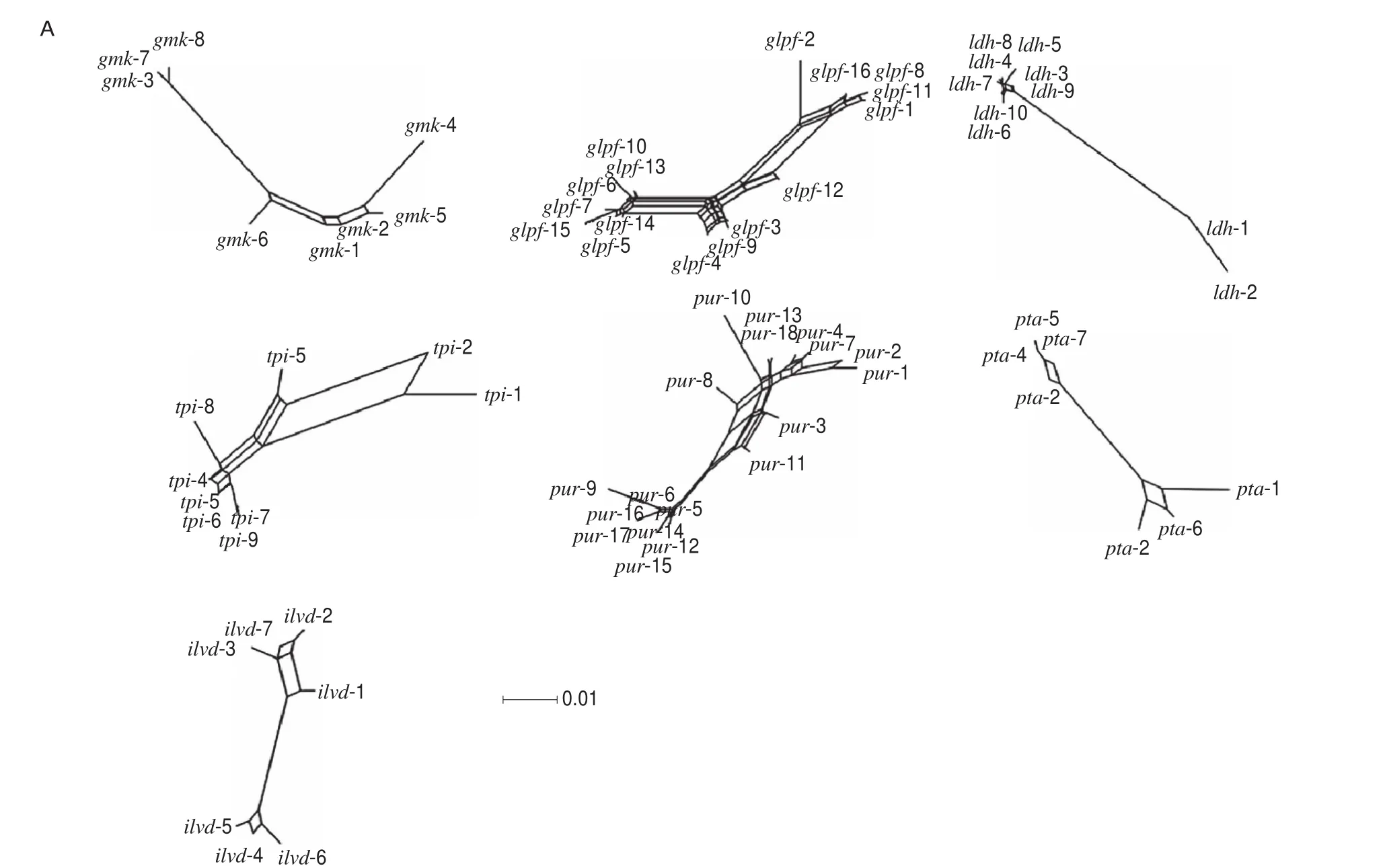
Fig. 1 Split decomposition analysis of B. coagulans. (A) Split decomposition analysis of B. coagulans based on 7 housekeeping gene fragments. (B) Split decomposition analysis of B. coagulans based on concatenated sequences of 7 housekeeping gene fragments.
3.1.3 Phylogenetic analysis of B. coagulans isolates
A minimum spanning tree was constructed to explore genetic lineages among theB. coagulansisolates according to geographical areas. All 33 STs were divided into 4 CCs and 7 singletons (Table S1,Fig. 2A, Fig. S2). A more detailed phylogenetic tree ofB. coagulansisolates was constructed based on 7 housekeeping genes. As shown in Fig. 2B, there were no direct relationships betweenB. coagulansisolates and isolation source or geographical areas.
3.2 Selection of B. coagulans strains for the animal trial
After the MLST analysis, 15 strains were randomly selected for further study according to the sequence type and the proximity relationships from the phylogenetic tree (Table S1, Fig. 2).Among these isolates, 5B. coagulansstrains (B. coagulans-1,B. coagulans-10,B. coagulans-39,B. coagulans-70 andB. coagulans-71)were selected for animal trials based onin vitrosporulation ability.Compared to other strains, these strains showed significantly higher spore yield (P< 0.05, Table 3). These isolates also showed high survival (60.77%–74.65%) and germination rate (33.8%–39.5%) in a simulated gastrointestinal tract (Table 4, Fig. 3).

Table 3Sporulation ability of 15 B. coagulans strains.

Table 4Survival rate of 5 B. coagulans strains in simulated gastrointestinal environment.
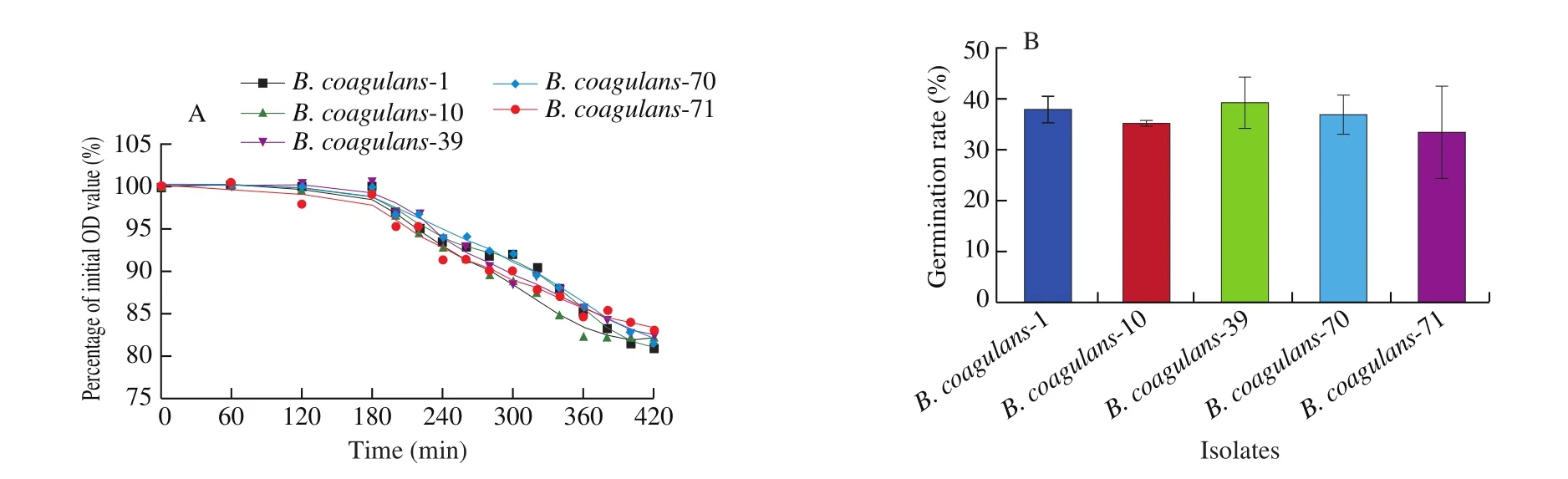
Fig. 3 Germination ability of B. coagulans in simulated gastrointestinal environment. (A) Germination curve of selected 5 strains of B. coagulans in simulated GI environment. (B) Germination rate of spores of 5 B. coagulans isolates.
3.3 Defecation status of mice
Compared to other test strains, the ingestion ofB. coagulans-39 significantly decreased indicators of constipation symptoms(e.g., increased number of fecal particles and weight of feces) in mice (P< 0.05, Fig. 4). In contrast, the fecal water content in theB. coagulans-10 group was similar to the constipation model group,suggesting that this strain had little effect on constipation.
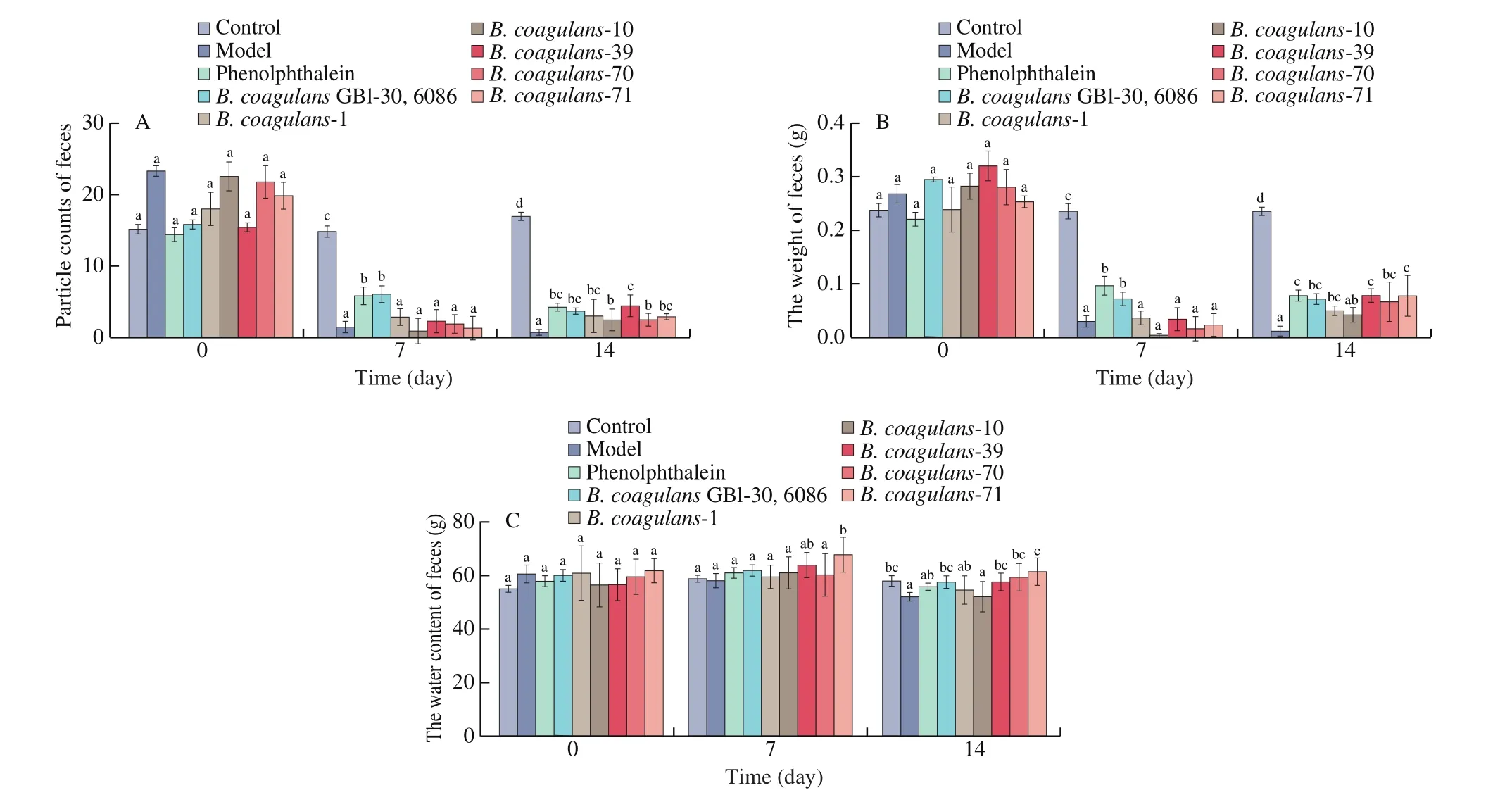
Fig. 4 Effects of B. coagulans on defecation status of mice. (A) The weight of feces. (B) The number of fecal particles. (C) The water content of feces. a-c There was a significant difference between the letters (P < 0.05).
3.4 Effect of B. coagulans on gastrointestinal transit
The time to the first black stool defecation in mice indicated that the ingestion ofB. coagulans-39 accelerated the peristalsis speed of mice intestines (P< 0.05, Fig. 5A). The defecation time in theB. coagulans-10 group was significantly longer than in the other treatment groups (P< 0.05). Similar results were observed for the small intestine transit rate. The small intestine transit rate in theB. coagulans-39 group was significantly higher compared to all the other groups (72.12%,P< 0.05), but the rate was lower in theB. coagulans-10 group (Fig. 5B).
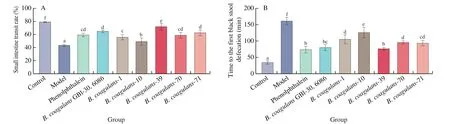
Fig. 5 Gastrointestinal transport capacity of mice. (A) Time to the first black stool defecation of mice. (B) Small intestine transit rate of mice. a-f There was a significant difference between the letters (P < 0.05).
3.5 Effect of B. coagulans isolates on SCFAs concentrations in feces
The SCFA concentrations in feces are shown in Table 5.Compared with the control group, the concentrations of acetic acid,propionic acid and total acid in the model group were significantly decreased (ANOVA,P< 0.05). On day 14, the levels of acetic acid, propionic acid, and total acid significantly increased in theB. coagulans-39,B. coagulans-70, andB. coagulans-71 groups compared with the model group (P< 0.05). After co-treatments with loperamide andB. coagulans-10, however, the levels of acetic acid,propionic acid and total acid were lower than in the model group.
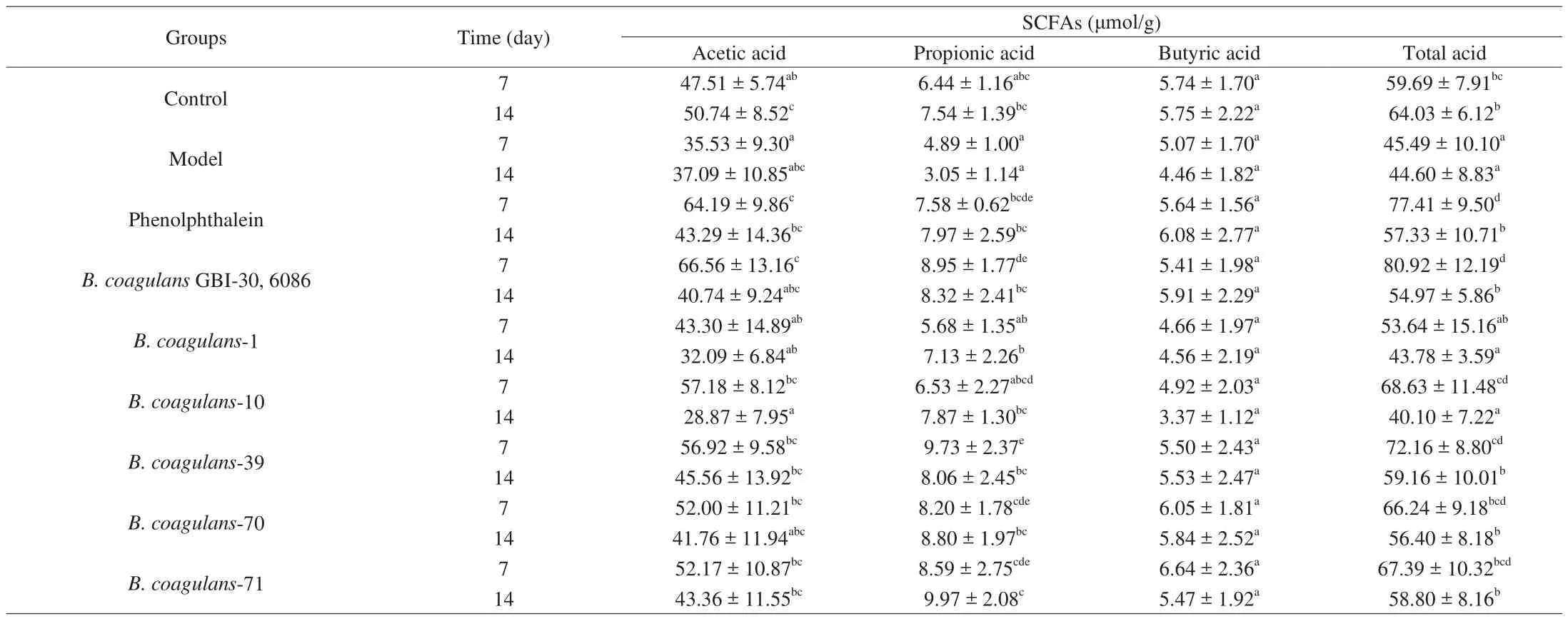
Table 5 Effects of different B. coagulans strains on the concentrations of SCFAs in feces.
3.6 Effect of B. coagulans isolates on fecal flora of constipated mice
B. coagulansstrains significantly increased the abundance and diversity of intestinal microbiota in constipated mice (Figs. 6 and 7).According to theβ-diversity analysis, microbiota composition was significantly altered in theB. coagulans-39,B. coagulans-70 andB. coagulans-71 groups (Fig. 6). After the ingestion ofB. coagulans-39, the relative abundance of some genera(e.g.,Bacillus,AllobaculuandDehalobacterium) were significantly up-regulated compared to other treatment groups(P< 0.05, Fig. 7).
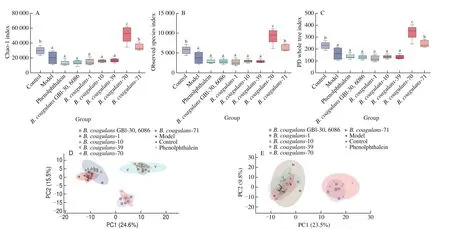
Fig. 6 Evaluation of the α-diversity and β-diversity in stool samples of mice. (A) Chao-1 index on 14th day. (B) Observed on 14th day Species index. (C) PD whole tree index on 14th day. (D) Principal component analysis of β-diversity in stool samples of mice on 7th day. (E) Principal component analysis of β-diversity in stool samples of mice on 14th day.
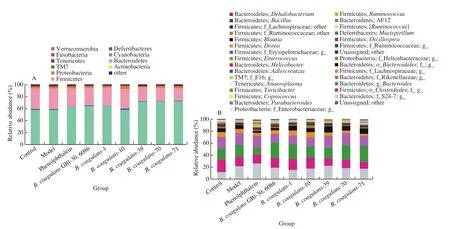
Fig. 7 The microbiota composition in feces of mice. Relative abundance of (A) main phyla in the different groups on 14th day, (B) main genera in the different groups on 14th day, (C) Bacillus, (D) Allobaculum, (E) Dehalobacterium, (F) Adlercreutzia. Different letters a-d indicate significant differences (P < 0.05).
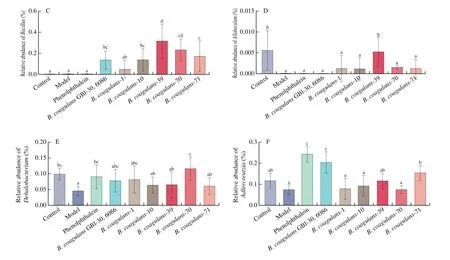
Fig. 7 (Continued)
3.7 Characterization of serum parameters of experimental mice
The effects ofB. coagulanson constipation were also evaluated by measurement of serum parameters. As shown in Table 6,supplementation with loperamide significantly decreased the level of motilin, gastrin and substance P and increased the level of somatostatin, vasoactive intestinal peptide, and endothelin 1 in mice.Compared with other test strains, the ingestion ofB. coagulans-39 andB. coagulansGBI-30, 6086 had more positive regulatory effects on the serum parameters (P< 0.05, Table 6).
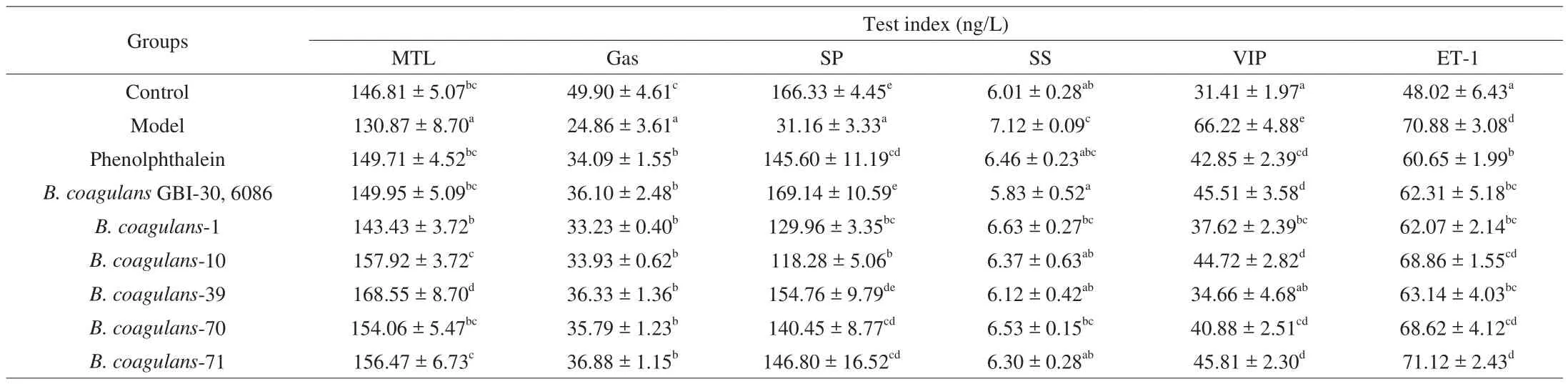
Table 6Effects of B. coagulans strains on serum parameters of mice.
3.8 Comparative genomic analysis of selected B. coagulans strains
The chromosomal features of the fiveB. coagulansstrains are shown in Table S3. The average nucleotide identity (ANI) values of whole genomes between homologous regions shared by every two genomes were generally above 97% (Table S4). A cluster of orthologous groups (COG) analysis showed a majority of the genes encoded proteins that were assigned to amino acid transport and metabolism (E), carbohydrate transport and metabolism (G)and function unknown (S) (Fig. 8A). A total of 20 COGs were observed inB. coagulans-39 when compared withB. coagulans-10(Table 7). Exploratory biological information on carbohydrate enzymes showed the endo-beta-1,4-galactanase gene related to GH53 was only detected inB. coagulans-39 (Fig. 8B). The generation time ofB. coagulans-39 (3.65 h) was significantly shorter than that ofB. coagulans-10 (5.71 h) in carbon-limited medium containing GOS (Fig. S3).
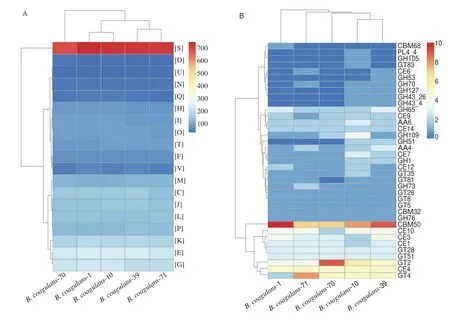
Fig. 8 The clusters of orthologous groups (COG) repertoires (A), and CAzyme families (B) of 5 B. coagulans strains.
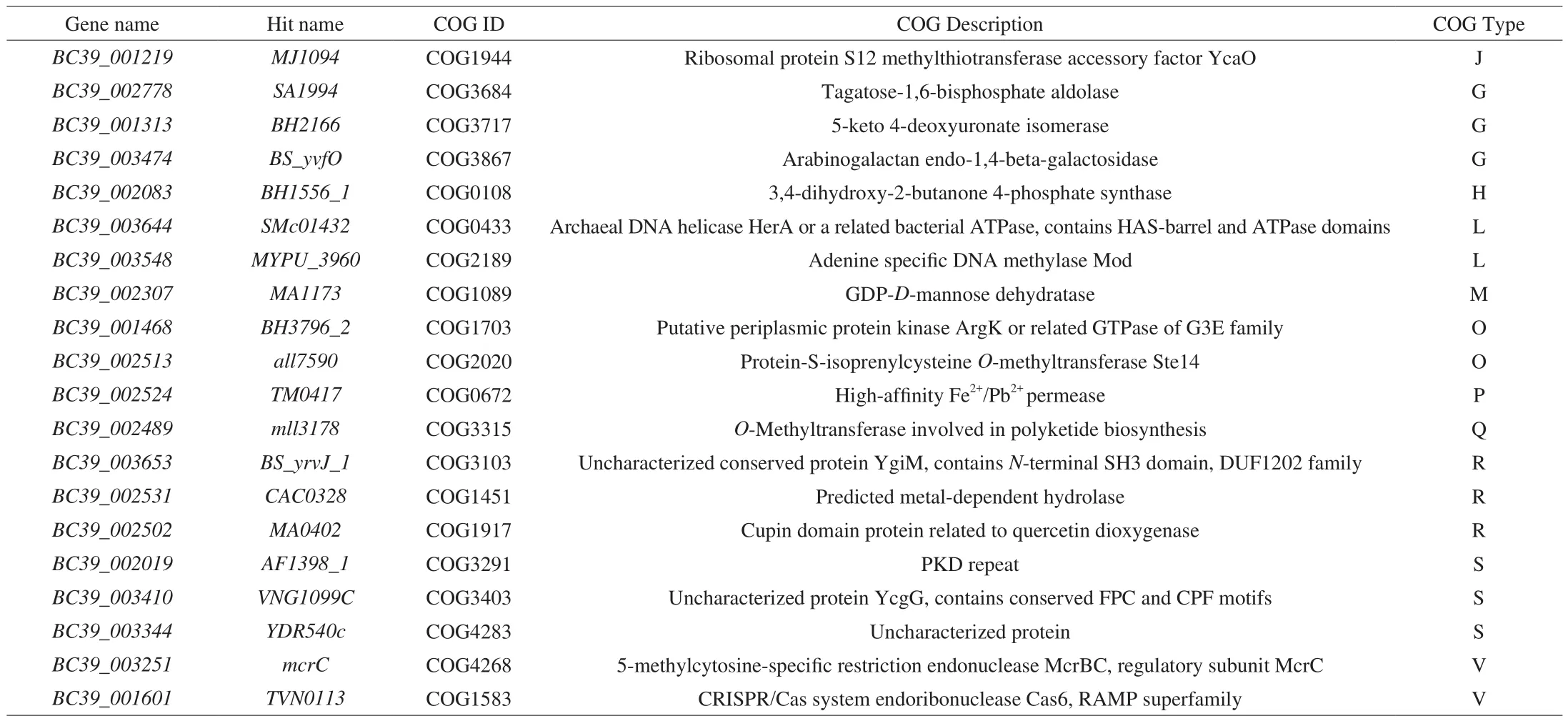
Table 7Strain-specific COG categories of B. coagulans-39 compared to B. coagulans-10.
4. Discussion
MLST analysis clearly showed intra-species heterogeneity amongB. coagulansstrains. The single nucleotide polymorphism value ofB. coagulans(6.92%) was higher than for other microbes in previous reports, suggesting the housekeeping genes ofB. coagulanshad high nucleotide genetic diversity and discrimination ability [38,39].Compared to other Bacillus species, such asB. cereus(Pivalue 0.019 9–0.085 6), the higherPivalue ofB. coagulans(0.016 38–0.041 45) confirmed the relatively high nucleotide diversity in this species [40]. The high nucleotide diversity of lactic acid bacteria(LAB) has been reported to be related to natural transformations and low frequency mutations [24]. The dN/dSratios ofB. coagulansin this study were all less than 1, indicating purifying selection of housekeeping genes against changes in amino acid composition, as has been previous reported for Bacillus licheniformis [41].
The evolutionary relationships betweenB. coagulansisolates and geography were also explored in this study. As shown in Fig. 2, the majority of the isolates from Eastern China were assigned to CC1.However,B. coagulansisolates from other geographical areas were widely separated in all CCs. Similar results have been found from MLST analysis ofC. difficile, supporting our conclusion that the evolution ofB. coagulanswas not correlated with geography [42]. In addition, there were no associations between STs and isolation source.Correlation analysis of the association between STs and geography(P> 0.05, Eta = 0.395) or isolation source (P> 0.05, Eta = 0.318)further confirmed the evolution direction ofB. coagulanswas not correlated with geography or isolation source (Table S3).
Our results also indicate thatB. coagulansisolates had a weak clonal population structure and strong indications of intraspecies recombination. Previous studies have shown that theIASofL. lactisandL. fermentumare 0.303 8 and 0.214 2, respectively [24,43].The lower value forIASofB. coagulansindicated the existence of a weaker clonal population structure in this species compared with traditional LAB isolates. Split graphs ofB. coagulansbased on concatenated sequences showed a complicated network structure with parallelogram-shaped structures, indicating recombination likely occurs frequently in this species and plays a role in generating genotypic diversity among the isolates. Similar results were reported in a previous study focusing onOenococcus oenistrains [38].
Published data have suggested that the integration of genotypic and phenotypic studies could significantly improve the relevance of screening strains adapted to specific functions and applications [44].Hence, 15 strains were randomly selected for study based on their genotype. To evaluate the function ofB. coagulansstrainsin vivo,several probiotic abilities were assessed. In general, the extreme environment of the human gastrointestinal tract greatly limits the growth of probiotics [45]. The indicators in this experiment thus ensured that the selected strains were able to adapt to the intestinal tract. These indicators guaranteed that spores of selectedB. coagulansstrains would probably germinate in the gastrointestinal tract. The 5B. coagulansstrains were also selected to evaluate their effects against constipation in mice.
The results from the animal trial indicated thatB. coagulansexerted a strain-specific effect on loperamide-induced constipation in mice. The analysis of feces parameters, intestinal transit ability,the content of SCFAs in feces, and microbiota flora in feces showed that supplementation ofB. coagulans-39 relieved constipation in mice. In contrast, the ingestion ofB. coagulans-10 had little effect against loperamide-induced constipation. The protective effects ofB. coagulansstrains against specific diseases (e.g., diarrhea and abdominal pain) differs or has even been invalidated because of strain-specific differences [46]. Similar results have also been found inLactobacillus salivariusandBifidobacterium adolescentis[21,47].
Finally, the potential genetic background amongB. coagulansisolates were explained by comparative genomic analysis. Twenty strain-specific COG categories were detected inB. coagulans-39 when compared toB. coagulans-10 (Table 7). Among the 20 genes assessed forB. coagulans-39, genesBC39_002778,BC39_001313,BC39_003474, andBC39_002307are responsible for carbohydrate uptake and degradation [48-50], geneBC39_00208is involved in the production of riboflavin [51], and geneBC39_002524is related to the metabolism of iron [52]. Some studies have reported that genes involved in the metabolism of rare sugars are closely related to the SCFAs production ability of probiotics [47]. For instance, supplementation of arabinogalactan in human fecal incubates increases the production of SCFAs [53]. Therefore, genesBC39_002778,BC39_001313, andBC39_003474detected inB.coagulans-39 may be associated with the highest SCFAs contents of the feces in theB. coagulans-39 group (Table 5). One animal trail indicated that microbially produced SCFAs in mice play an important role in modulating small intestinal transit [54]. Based on these analyses, compared to other strains, the higher content of SCFAs detected in theB. coagulans-39 group may be an important factor leading to the higher small intestine transit rate in this group so as to relive the symptoms of constipation in mice (Fig. 5).COG (Genes:BC39_002778,BC39_001313,BC39_003474, andBC39_002307) and CAZyme (Genes related to GH53) analysis in the present study suggested thatB. coagulans-39 could use various carbohydrates (e.g., GOS and arabinogalactan) in the gastrointestinal tract [36]. Furthermore, ribo flavin encoded by geneBC39_00208is also an essential water-soluble vitamin that acts as the coenzyme in metabolic pathways for carbohydrate and protein metabolism [51].A previous study reported that the adaptability of probiotics to nutrients and environmental conditions in the gastrointestinal tract is important to their residence time and survival rate [55]. In agreement with this finding, supplementation withB. coagulans-39 induced a higher abundance ofBacillusthan the other test strains (P< 0.05,Fig. 5D). Supporting to these results, our studies indicated thatB.coagulans-39 may be more suitable for the intestinal tract of mammal and may have higher viable bacteria count in the gut compared to otherB. coagulansstrains. Similar to other probiotic strains (e.g.B. adolescentis), the high viable bacteria count ofB. coagulans-39 exhibited strain-specific effects in the alleviation of constipation due to its effects on the gut microbiome and microenvironment [21].Moreover, geneBC39_002524encoding high-affinity Fe2+/Pb2+permease is closely related to the metabolism of iron thereby reducing the risk of constipation. It is generally recognized that gastrointestinal disturbance, particularly constipation, is a common side effect caused by the supplement of iron [56]. Our results further confirmed the integration of genotypic and phenotypic characteristics of strains may be an effective method to screen strains adapted to specific functions,which were consistent with the results of previous studies [44,47].
5. Conclusions
The present study provides a better understanding of the genetic relationships among 57 isolates ofB. coagulansusing MLST.B. coagulansexerted a strain-specific effect on loperamide-induced constipation in mice. Among the strains selected in this study,B. coagulans-39 showed a better ability to alleviate constipation.Comparative genome analysis detected a series of carbohydrate utilization genes inB. coagulans-39, which meansB. coagulans-39 might proliferate in the gastrointestinal environment using various carbohydrates as primary substrates. This ability guarantees a sufficient number ofB. coagulans-39 cells in the gastrointestinal tract,thereby likely contributing to more protection against constipation in mice compared to otherB. coagulansstrains.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China Program [No. 31871773 and No. 31820103010];Projects of Innovation and Development Pillar Program for Key Industries in Southern Xinjiang of Xinjiang Production and Construction Corps [2018DB002]; National First-Class Discipline Program of Food Science and Technology [JUFSTR20180102];the BBSRC Newton Fund Joint Centre Award; and Collaborative Innovation Center of Food Safety and Quality Control in Jiangsu Province.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://doi.org/10.1016/j.fshw.2022.03.014.
杂志排行
食品科学与人类健康(英文)的其它文章
- Dietary bioactives and essential oils of lemon and lime fruits
- Green tea, epigallocatechin gallate and the prevention of Alzheimer’s disease: clinical evidence
- Simultaneous quantification of 18 bioactive constituents in Ziziphus jujuba fruits by HPLC coupled with a chemometric method
- A systematic study on mycochemical profiles, antioxidant, and anti-inflammatory activities of 30 varieties of Jew’s ear (Auricularia auricula-judae)
- GPP (composition of Ganoderma lucidum polysaccharides and Polyporus umbellatus polysaccharides) protects against DSS-induced murine colitis by enhancing immune function and regulating intestinal flora
- Immunoregulatory polysaccharides from Apocynum venetum L.flowers stimulate phagocytosis and cytokine expression via activating the NF-κB/MAPK signaling pathways in RAW264.7 cells
