拟除虫菊酯类农药的微生物降解及其机制研究进展
2021-09-26赵叶子陈智坤王铮吴桐张颖萍徐倩
赵叶子 陈智坤 王铮 吴桐 张颖萍 徐倩


摘要:拟除虫菊酯被认为是有机磷农药的安全替代品,因此当有机磷农药被禁限使用时,其应用显著增加。目前,拟除虫菊酯销量约占世界杀虫剂总额的20%。这类农药的长期、广泛使用既带来了经济效益,也造成了环境污染,危害人类及其他非靶标生物。针对这一问题,已开发出多项修复技术,其中微生物法高效环保、成本低廉,已成为修复拟除虫菊酯污染的最优方法。笔者综述了最新分离的拟除虫菊酯降解菌株及其特性;拟除虫菊酯降解酶及其基因;拟除虫菊酯及其代谢产物(间苯氧基苯甲酸等)的降解途径。此外,还提出了拟除虫菊酯类农药微生物降解研究的发展趋势和需要进一步解决的问题。
关键词:拟除虫菊酯;微生物降解;降解酶;降解途径;间苯氧基苯甲酸
中图分类号:X592 文献标志码:A
文章编号:1002-1302(2021)17-0028-11
收稿日期:2021-02-01
基金项目:西安市科技局项目(编号:2017127SF/SF021);。
作者简介:赵叶子(1992—),女,陕西西安人,硕士研究生,主要从事微生物有机污染修复研究。E-mail:jzzszyj8943@163.com。
通信作者:王 铮,博士,副教授,主要从事环境评价与水处理研究。E-mail:Wang_zheng1992@126.com。
拟除虫菊酯类农药是人工合成的天然除虫菊酯的类似物[1],根据拟除虫菊酯分子中是否含有氰基,将其分为Ⅰ型和Ⅱ型,Ⅱ型具有氰基及更强的毒性[2,3]。
拟除虫菊酯通过开放钠离子通道,实现杀虫目的[4]。因其高效的杀虫能力,使其在农林业生产中发挥重要作用[5]。随着该类农药的大范围大量使用,其负面影响被不断报道。拟除虫菊酯及其代谢产物在环境及人体尿液、母乳中普遍检出,其对人体神经系统、生殖系统造成干扰,可能引起血液癌症、氧化应激及DNA损伤等危害已被证实[6-11]。因此,世界各国都制定了农产品中拟除虫菊酯类农药残留的限定标准[12-13]。
针对此环境问题,微生物降解是一种环境及经济友好的修复技术,目前研究者已在实验室获得大量研究成果,对这些研究成果的梳理,将有助于农药微生物降解技术的进一步发展和应用。
本文综述了已分离的拟除虫菊酯降解菌株,及其生理生化特性、关键功能基因和酶、拟除虫菊酯降解途径,并重点关注了其代谢产物间苯氧基苯甲酸(3-phenoxybenzoic acid,简称PBA)等的降解,以期为拟除虫菊酯的安全利用与污染修复提供参考。
1 降解菌株及其生理生化、降解特性
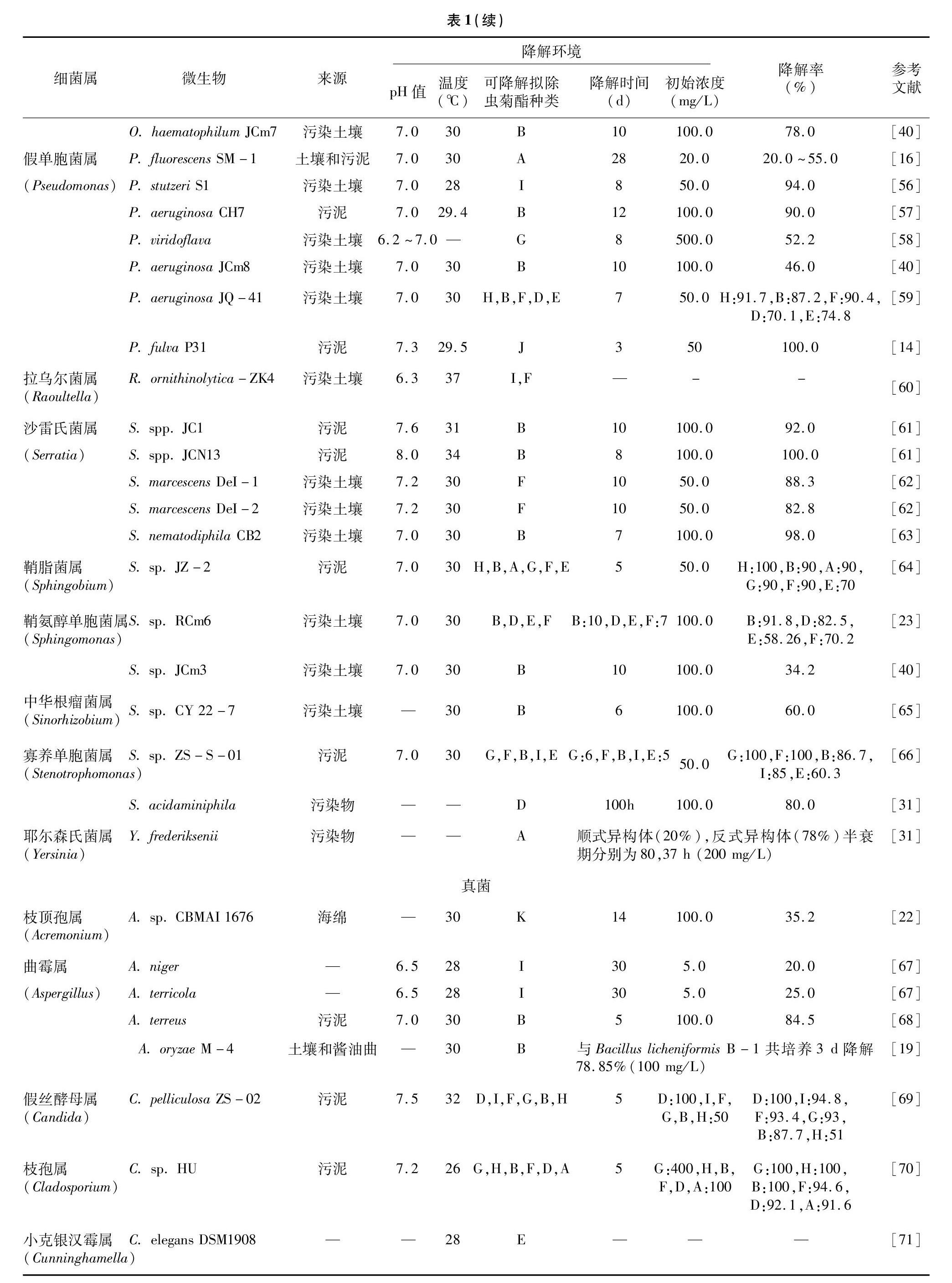
微生物可将有机污染物降解成更具安全性的产物,甚至完全矿化,广泛应用于环境污染物的降解中[10,14]。Kaufman等最初描述了苄氯菊酯在土壤中的降解[15]。1988年Maloney等从来自污水处理厂的样品中首次得到了能够有效降解拟除虫菊酯的菌株,共得到3个菌株,分别为荧光假单胞菌(Pseudomonas fluorescens) SM-1、无色杆菌属(Achromobacter sp.) SM-2、蜡样芽孢杆菌(Bacillus cereus) SM-3[16]。自此,大量可以有效降解拟除虫菊酯的菌种被筛选鉴定。这些菌种大都能以某种拟除虫菊酯作为唯一碳源,进行生长代谢。笔者共整理了来自45个不同菌属的拟除虫菊酯降解菌,包括细菌26个菌属、真菌14个菌属、放线菌5个菌属,其生理生化、降解特性等详细信息见表1。所有菌属中,芽孢杆菌属(Bacillus)、苍白杆菌属(Ochrobactrum)、假单胞菌属(Pseudomonas)、沙雷氏菌属(Serratia)及链霉菌属(Streptomyces)包含了较多菌株。大部分功能菌分离自污染环境,如污染土壤、污水、活性污泥等,亦有以茶葉、酒曲、动植物体,甚至来自大西洋的海绵等作为分离源的报道[17-22]。绝大多数菌株在温和环境(pH值=7,30 ℃ 左右)中有较高活性。受益于拟除虫菊酯结构的相似性,筛选所得的菌株,往往具有对拟除虫菊酯类农药降解的广谱性,如乙酸钙不动杆菌(Acinetobacter calcoaceticus) MCm5[23]、类短短芽孢杆菌(Brevibacillus parabrevis) FCm9[23]、小链小杆菌属(Catellibacterium sp.) CC-5[24]等。
具有广谱、高效、高耐受性、降解彻底特性的菌株是研究者期待获得的菌株。虞云龙等分离得到具有农药降解广谱性的菌株产碱菌属(Alcaligenes) sp. YF11,能够同时降解部分拟除虫菊酯和一硫代磷酸酯[25]。陈锐等筛选出的草酸青霉(Penicillium oxalicum) SSCL-5,可在24 h内降解97%的氯氰菊酯(初始浓度400 mg/L),具有较为突出的降解速率[26]。Chen等从土壤中分离出的Bacillus sp. DG-02,可耐受浓度高达1 200 mg/L的甲氰菊酯,能有效降解7种拟除虫菊酯,并在土壤修复中取得较好的效果,证明了其实际应用潜力[27],该研究组之后分离得到的菌株苏云金芽孢杆菌(Bacillus thuringiensis) ZS-19[28]、黄褐假单胞菌(Pseudomonas fulva) P31[14],亦有广谱、耐受性强及降解效果好的特点,其中Bacillus thuringiensis ZS-19 对于PBA具有很好的降解效果[28]。研究者通过构建复合菌系协同降解的方式使矿化更为彻底,Zhao等将地衣芽孢杆菌(Bacillus licheniformis) B-1 和米曲霉(Aspergillus oryzae) M-4共同培养用于氯氰菊酯的降解[19],相较单一菌种的降解,提高了矿化程度。
拟除虫菊酯具有1~3个手型中心,2~8个立体异构体[2-3],因此,菌株对于拟除虫菊酯的降解显示出结构选择性[29-30],Lee等发现菌株弗氏耶尔森菌(Yersinia frederiksenii)、温和气单胞菌(Aeromonas sobria)、 Erwinia carotovora在降解苄氯菊酯过程中,反式异构体相对顺式异构体更易被降解[31]。
除去传统的富集培养筛选法,研究者也通过构建基因工程菌的方式获取菌株。Lan等设计载体pETDuet,用于同时表达2个目的基因。在共表达载体上克隆了黄杆菌属(Flavobacterium sp.)中的有机磷水解酶基因opd和来自尖音库蚊(Culex pipiens)的酯酶基因B1,实现了单一微生物同时降解有机磷、氨基甲酸酯和拟除虫菊酯杀虫剂的目的[32],为复配农药的同时降解提供了新思路。
目前,所取得的成果多局限于实验室,在实际应用方面还缺少报道。构建具有优越特性的基因工程菌是值得探索的研究方向。
2 降解拟除虫菊酯的酶及基因
2.1 降解酶
在微生物降解有机污染物过程中,实际起作用的是微生物体内的各种酶。降解酶往往比产生这种酶的微生物细胞具有更强的抗逆性及降解效率,特别是在低农药浓度条件下[77]。早在1993年,Maloney等通过层析技术从Bcillus cereus SM3中分离获得1种羧酸酯酶,命名为氯菊酯酶(permethrinase ),并采用离子交换色谱法和凝胶过滤色谱法进行纯化,首次实现了通过无细胞的酶系统降解拟除虫菊酯[78]。类似的,从黑曲霉(Aspergillus niger) ZD11细胞提取物中提取纯化的新型拟除虫菊酯水解酶,分子量为56 ku、pI值(等电点)为5.4,当温度为45 ℃、pH值为6.5时酶活性最佳,该酶优先降解对于哺乳动物更具毒害作用的反式异构体拟除虫菊酯,人肝脏羧酸酯酶也存在这一特性[79-80]。1993年至今,陆续有研究者对拟除虫菊酯降解酶进行纯化(部分酶信息见表2),多为酯酶。其中大多数酶为胞内酶,而铜绿假单胞菌(Pseudomonas aeruginosa) GF31降解氯氰菊酯过程中起催化作用的氨肽酶是一种胞外酶[81]。图1描述了表2所列部分酶之间的进化关系(作图软件为MEGA X 64-bit,蛋白质序列来自:https://www.ncbi.nlm.nih.gov/protein)。2018年,Gangola等发现漆酶参与了氯氰菊酯降解过程,这是首次发现漆酶作用于拟除虫菊酯降解[45]。漆酶已被发现可用于多种有机污染的环境修复,白腐菌是常见的漆酶生产源,其在食用菌菌渣中也大量存在[82]。这无疑为菌渣的利用提供了新途径。
2.2 降解基因
随着基因工程技术的进步,对于拟除虫菊酯降解酶的研究已发展到基因层面。2006年,Wu等通过构建基因文库,从Klebsiella sp. ZD112中得到拟除虫菊酯水解酶基因EstP,基因全长1 924 bp,编码637个氨基酸,该酯酶不但可以降解拟除虫菊酯,还可以催化降解其他底物,如有机磷农药,这是关于拟除虫菊酯降解基因的首次报道[80]。Hu等从拟除虫菊酯降解菌Bacillus cereus BCC01的基因组文库中筛选并鉴定了关键的拟除虫菊酯水解酶基因EstA,该酶的Ser94位于固定的五肽基序列Gly-X-Ser-X-Gly中,形成了一个凹形的活性中心,用于氯氰菊酯的生物降解,这是酯酶的典型特征[46]。丝氨酸在酶的催化位点参与酰化反应具有类似的情况,已在几种拟除虫菊酯水解酶中发现,如Sys410[86]、PytY[87]、Pye3[83]和Est684[91]。
随着基因层面研究的深入,宏基因组技术作为一种辅助工具帮助研究者更具方向性地达到科研目的。Hong等将甲基对硫磷降解基因mpd引入能够降解甲氰菊酯的菌株Sphingobium sp. JQL4-5的染色体中,成功构建出能够同时降解2种污染物的菌株[93]。目前此类研究成果相对有限,是未来期待发展的方向。
3 降解途径
在不同报道中,拟除虫菊酯降解途径有相似性,但在中间过程及产物方面存在差异,这些差异可能是由于微生物的种类、拟除虫菊酯的结构及培养条件差异造成的。
3.1 酯键的断裂
拟除虫菊酯微生物降解机制的核心部分是酯键的断裂,将原农药分解为羧酸和醇[44]。此步骤通常为降解过程的的第1步,以Ochrobactrum tritici pyd-1降解甲氰菊酯为例,酯键断裂后甲氰菊酯分解为间苯氧基氰基苄醇(3-phenoxybenzyl alcohol,PBAlc)和2,2,3,3-四甲基环丙烷甲酸(图2)[54]。类似过程中,多数情况下含环丙烷基团的降解产物为酸,含间苯氧基基团的产物为醇。也存在特殊情况,如在Brevibacterium aureum降解三氟氯氰菊酯过程中,原农药首先被分解为2,2,3,3-四甲基环丙烷甲醇和4-氟代-3-苯氧基苯甲酸(图3)[48]。
大多数微生物对拟除虫菊酯的降解遵循上述规律,也存在特例,如Lysinibacillus sphaericus FLQ-11-1降解氟氯氰菊酯过程中,第1步并非打开酯键[52]。
3.2 常见代谢产物及其转化关系
“3.1”节相关降解案例中出现的PBAlc,以及间苯氧基苄醛(3-Phenoxybenzaldehyde,PBAld)和PBA是拟除虫菊酯降解过程中最常见的代谢物,相比母体它们具有更强的毒性[94-95]。如PBA半衰期长达180 d[96],具有雌性激素特性,是一种内分泌米干扰物,对人体具有一定毒性[97-98]。
大多数拟除虫菊酯降解过程中都会出现这些常见的中间产物,特别是结构中包括间苯氧基基团的拟除虫菊酯,而氟氯氰菊酯、联苯菊酯、丙烯菊酯等不包括典型间苯氧基基团的拟除虫菊酯在降解过程中生成其他产物,但依然遵循酯键断裂生成醇和酸的规律。如Acidomonas sp. 降解烯丙菊酯过程中生成一个烯丙酮醇[33],联苯菊酯的微生物降解过程中,检测到2-甲基-3-联苯甲醇[69]。
在拟除虫菊酯代谢过程中,PBAlc、PBAld及PBA三者之间常常相互转化,Bacillus licheniformis CY-012[99]、Pseudomonas fluorescens SM-1[16]可氧化PBAlc为PBA,Acinetobacter baumannii ZH-14[34]转化PBAlc为PBAld,进一步氧化为PBA。而Microsphaeropsis sp. CBMAI1675[22]可将PBAld轉化为PBAlc,在Aspergillus oryzae M-4[100]降解目标物过程中,可观察到PBAlc与PBA的相互转换。
3.3 常见代谢产物的降解途径
相比拟除虫菊酯本身,针对其代谢产物如何降解的研究较少,但此类研究同样具有重要意义。其中,被讨论较多的代谢产物是PBA[10]。
早期的相关研究仅限于细菌范畴,特别是Pseudomonas sp.菌株。1990年Engesser等首次分离得到1株不能完全降解PBA的菌株类产碱假单胞菌(Pseudomonas pseudoalcaligenes) POB310[101],并在后续研究中构建了基因工程菌Pseudomonas sp. B13-D5及Pseudomonas sp. B13-ST1,新菌株同时具有降解氯酚的能力[96]。亦有研究者进行了Micrococcus sp.[53]、Sphingobium sp.[64,84]、Bacillus sp.[27]、Sphingobium sp.[39,102]及Stenotrophomonas sp.[66]等细菌菌属针对PBA降解的研究。
针对PBA降解的真菌领域研究起步较晚。首次报道是在2012年,袁怀瑜等筛选得到可在22 h内完全降解100 mg/L PBA的菌株Aspergillus niger YAT1[103-104]。Eurotium cristatum ET1亦为PBA的有效降解真菌菌株,其代谢产物包括苯酚和邻苯二酚[17]。Aspergillus oryzae M-4降解PBA的产物食子酸为有毒代谢物[100],该菌株无法单独代谢,加入Bacillus licheniformis B-1共培养可降解没食子酸,实现无毒化[19],真菌中常有的木质素降解酶漆酶、LiP和MnP也与PBA的降解相关[94],它们在食用菌菌渣中被普遍检测到[82]。
综上所述,并非所有的拟除虫菊酯降解研究均实现了完全矿化、无毒化。构建复合菌系可以帮助实现这一目标。
4 结论与展望
隨着世界范围内人们环保意识的增强,研究者们就微生物修复农药污染开展了大量研究,其中关于拟除虫菊酯的降解也取得了较好的研究成果。笔者主要从降解菌株、降解基因及酶、降解机制几个方面对拟除虫菊酯类农药的微生物降解的现有研究进行了综述。首次对涉及45个菌属的拟除虫菊酯降解菌株信息进行了总结归纳;对拟除虫菊酯的降解基因及酶进行了信息总结,着重降解特性及分类;通过对微生物降解拟除虫菊酯的不同案例进行归纳整理,总结出了拟除虫菊酯降解途径的一般性规律,及可能存在的特殊情况;并着重分析了代谢产物PBA的降解,较为全面地对1990年至今的相关研究成果进行了总结。
现有研究成果为将来拟除虫菊酯类农药的降解及微生物环境修复相关研究提供了思路:(1)现有的大量研究成果,为利用基因工程技术,人为构建更为高效、广谱、抗逆的基因工程菌提供了良好基础;(2)构建多功能复合菌系,是以较低成本实现高效矿化有机污染物的途径,现有微生物降解拟除虫菊酯研究极少涉及这一领域;(3)土壤环境降解的研究有利于克服实际应用过程中的复杂环境因素,现有研究在土壤环境降解研究方面较为薄弱;(4)拟除虫菊酯的常见代谢产物PBA等,具有相比原农药更强的毒性及半衰期,但针对性的环境修复研究较少,此类研究具有重要意义;(5)随着萃取技术的优化,低抗性、易降解的天然除虫菊酯大量应用具有了可行性[105],推进此过程或开发更为高效、环保的新型菊酯,并进行相关微生物降解研究,亦有利于降低拟除虫菊酯造成的环境负担。
参考文献:
[1]Smith T M,Stratton G W. Effects of synthetic pyrethroid insecticides on nontarget organisms[J]. Residue Reviews,1986,97:93-120.
[2]Kaviraj A,Gupta A. Biomarkers of type Ⅱ synthetic pyrethroid pesticides in freshwater fish[J]. BioMed Research International,2014(9):928063.
[3]Laskowski D A. Physical and chemical properties of pyrethroids[J]. Reviews of Environmental Contamination and Toxicology,2002,174:49-170.
[4]Chrustek A,Holyńska-Iwan I,Dziembowska I,et al. Current research on the safety of pyrethroids used as insecticides[J]. Medicina,2018,54(4):61.
[5]Kuivila K M,Hladik M L,Ingersoll C G,et al. Occurrence and potential sources of pyrethroid insecticides in stream sediments from seven U.S. metropolitan areas[J]. Environmental Science & Technology,2012,46(8):4297-4303.
[6]Gammon D W,Liu Z,Chandrasekaran A,et al. Pyrethroid neurotoxicity studies with bifenthrin indicate a mixed type Ⅰ/Ⅱ mode of action[J]. Pest Management Science,2019,75(4):1190-1197.
[7]Ye X Q,Pan W Y,Zhao S L,et al. Relationships of pyrethroid exposure with gonadotropin levels and pubertal development in Chinese Boys[J]. Environmental Science & Technology,2017,51(11):6379-6386.
[8]Navarrete-Meneses M,Pérez-Vera P. Pyrethroid pesticide exposure and hematological cancer:epidemiological,biological and molecular evidence[J]. Reviews on Environmental Health,2019,34(2):197-210.
[9]Zepeda-Arce R,Rojas-García A E,Benitez-Trinidad A,et al. Oxidative stress and genetic damage among workers exposed primarily to organophosphate and pyrethroid pesticides[J]. Environmental Toxicology,2017,32(6):1754-1764.
[10]Huang Y C,Xiao L J,Li F Y,et al. Microbial degradation of pesticide residues and an emphasis on the degradation of cypermethrin and 3-phenoxy benzoic acid:a review[J]. Molecules,2018,23(9):2313.
[11]夏曉华,张林霞,司松波,等. 高效氯氟氰菊酯对泥鳅的急性毒性及生理毒性[J]. 江苏农业科学,2013,41(6):270-272.
[12]国家卫生健康委员会,中华人民共和国农业农村部,国家市场监督管理总局. 食品安全国家标准 食品中农药最大残留限量:GB 2763—2019[S]. 北京:中国标准出版社,2019.
[13]EU. Amending annexes Ⅱ,Ⅲ and Ⅴ to regulation (EC):396/2005[S]. EU,2005.
[14]Yang J J,Feng Y M,Zhan H,et al. Characterization of a pyrethroid-degrading Pseudomonas fulva strain P31 and biochemical degradation pathway of d-phenothrin[J]. Frontiers in Microbiology,2018,9:1003.
[15]Kaufman D D,Haynes S C,Jordan E G,et al. Permethrin degradation in soil and microbial cultures[J]. American Chemical Society,1977,42:147-161.
[16]Maloney S E,Maule A,Smith A R. Microbial transformation of the pyrethroid insecticides:permethrin,deltamethrin,fastac,fenvalerate,and fluvalinate[J]. Applied and Environmental Microbiology,1988,54(11):2874-2876.
[17]Hu K D,Deng W Q,Zhu Y T,et al. Simultaneous degradation of β-cypermethrin and 3-phenoxybenzoic acid by Eurotium cristatum ET1,a novel “golden flower fungus” strain isolated from Fu Brick Tea[J]. Microbiology Open,2018,8(7):e776.
[18]孙丽娜,黄开华,高新华,等. 具氯氰菊酯降解功能的植物内生细菌分离鉴定及降解特性研究[J]. 农业环境科学学报,2020,39(1):70-77.
[19]Zhao J Y,Chi Y L,Xu Y C,et al. Co-metabolic degradation of β-cypermethrin and 3-phenoxybenzoic acid by co-culture of Bacillus licheniformis B-1 and Aspergillus oryzae M-4[J]. PLoS One,2016,11(11):e0166796.
[20]Tian J W,Long X F,Zhang S,et al. Screening cyhalothrin degradation strains from locust epiphytic bacteria and studying Paracoccus acridae SCU-M53 cyhalothrin degradation process[J]. Environmental Science and Pollution Research International,2018,25(12):11505-11515.
[21]王若瑜,曹 明,陈晶瑜.大曲中氯氟氰菊酯降解菌的分离筛选及特性研究[J]. 中国酿造,2020,39(3):21-25.
[22]Birolli W G,Alvarenga N,Seleghim M H,et al. Biodegradation of the pyrethroid pesticide esfenvalerate by Marine-Derived fungi[J]. Marine Biotechnology,2016,18(4):511-520.
[23]Akbar S,Sultan S,Kertesz M. Determination of cypermethrin degradation potential of soil bacteria along with plant growth-promoting characteristics[J]. Current Microbiology,2015,70(1):75-84.
[24]Zhao H,Geng Y,Chen L,et al. Biodegradation of cypermethrin by a novel Catellibacterium sp. strain CC-5 isolated from contaminated soil[J]. Canadian Journal of Microbiology,2013,59(5):311-317.
[25]虞云龍,宋凤鸣,郑 重,等. 一株广谱性农药降解菌(Alcaligenes sp.)的分离与鉴定[J]. 浙江农业大学学报,1997(2):3-7.
[26]陈 锐,瞿 佳,孙晓宇,等. 氯氰菊酯降解菌草酸青霉SSCL-5分离鉴定及降解特性[J]. 生物技术通报,2020,36(6):120-127.
[27]Chen S H,Deng Y Y,Chang C Q,et al. Fenpropathrin biodegradation pathway in Bacillus sp. DG-02 and its potential for bioremediation of pyrethroid-contaminated soils[J]. Journal of Agricultural and Food Chemistry,2014,62(10):2147-2157.
[28]Chen S H,Deng Y Y,Chang C Q,et al. Pathway and kinetics of cyhalothrin biodegradation by Bacillus thuringiensis strain ZS-19[J]. Scientific Reports,2015,5(1):8784.
[29]Pérez-Fernández V,García M A,Marina M L. Characteristics and enantiomeric analysis of chiral pyrethroids[J]. Journal of Chromatography A,2010,1217(7):968-989.
[30]张亚杰,李劭彤,李朝阳,等. 菊酯农药微生物降解的异构体选择性特征[J]. 江苏农业科学,2019,47(10):278-280,290.
[31]Lee S,Gan J Y,Kim J S,et al. Microbial transformation of pyrethroid insecticides in aqueous and sediment phases[J]. Environmental Toxicology and Chemistry,2004,23(1):1-6.
[32]Lan W S,Gu J D,Zhang J L,et al. Coexpression of two detoxifying pesticide-degrading enzymes in a genetically engineered bacterium[J]. International Biodeterioration & Biodegradation,2006,58(2):70-76.
[33]Paingankar M,Jain M,Deobagkar D. Biodegradation of allethrin,a pyrethroid insecticide,by an Acidomonas sp.[J]. Biotechnology Letters,2005,27(23/24):1909-1913.
[34]Zhan H,Wang H S,Liao L S,et al. Kinetics and novel degradation pathway of permethrin in Acinetobacter baumannii ZH-14[J]. Frontiers in Microbiology,2018,9(2):98.
[35]Tang J,Hu Q,Lei D,et al. Characterization of deltamethrin degradation and metabolic pathway by co-culture of Acinetobacter junii LH-1-1 and Klebsiella pneumoniae BPBA052[J]. AMB Express,2020,10(1):106.
[36]Ma Y,Chen L S,Qiu J G. Biodegradation of beta-cypermethrin by a novel Azoarcus indigens strain HZ5[J]. Journal of Environmental Science and Health,2013,48(10):851-859.
[37]黄文文,刘晶晶,叶美玲,等. 甲氰菊酯降解菌Z1-3的分离鉴定及讲解特性[J]. 西北农林科技大学学报(自然科学版),2010(10):103-109,114.
[38]Sundaram S,Das M T,Thakur I S. Biodegradation of cypermethrin by Bacillus sp.in soil microcosm and in-vitro toxicity evaluation on human cell line[J]. International Biodeterioration & Biodegradation,2013,77(1):39-44.
[39]Liu F F,Chi Y L,Wu S,et al. Simultaneous degradation of cypermethrin and its metabolite,3-phenoxybenzoic acid,by the cooperation of Bacillus licheniformis B-1 and Sphingomonas sp. SC-1[J]. Journal of Agricultural and Food Chemistry,2014,62(33):8256-8262.
[40]Akbar S,Sultan S,Kertesz M. Bacterial community analysis of cypermethrin enrichment cultures and bioremediation of cypermethrin contaminated soils[J]. Journal of Basic Microbiology,2015,55(7):819-829.
[41]Tiwary M,Dubey A K. Cypermethrin bioremediation in presence of heavy metals by a novel heavy metal tolerant strain,Bacillus sp. AKD1[J]. International Biodeterioration & Biodegradation,2016,108(3):42-47.
[42]Panka J,Sharma A,Gangola S,et al. Novel pathway of cypermethrin biodegradation in a Bacillus sp. strain SG2 isolated from cypermethrin-contaminated agriculture field[J]. 3 Biotech,2016,6(1):45.
[43]Lee Y S,Lee J H,Hwang E J,et al. Characterization of biological degradation cypermethrin by Bacillus amyloliquefaciens AP01[J]. Journal of Applied Biological Chemistry,2016,59(1):9-12.
[44]苏宏南,陈切希,赵甲元,等. 微生物共培养降解β-氯氰菊酯的适宜条件[J]. 食品与发酵工业,2018,44(7):8-12.
[45]Gangola S,Sharma A,Bhatt P,et al. Presence of esterase and laccase in Bacillus subtilis facilitates biodegradation and detoxification of cypermethrin[J]. Scientific Reports,2018,8(1):12755.
[46]Hu W,Lu Q Q,Zhong G H,et al. Biodegradation of pyrethroids by a hydrolyzing carboxylesterase EstA from Bacillus cereus BCC01[J]. Applied Sciences-Basel,2019,9(3):e414-e477.
[47]Tang J,Liu B,Chen T T,et al. Screening of a beta-cypermethrin-degrading bacterial strain Brevibacillus parabrevis BCP-09 and its biochemical degradation pathway[J]. Biodegradation,2018,29(6):525-541.
[48]Chen S H,Dong Y H,Chang C Q,et al. Characterization of a novel cyfluthrin-degrading bacterial strain Brevibacterium aureum and its biochemical degradation pathway[J]. Bioresource Technology,2013,132(1):16-23.
[49]Zhang S B,Yin L B,Liu Y,et al. Cometabolic biotransformation of fenpropathrin by Clostridium species strain ZP3[J]. Biodegradation,2011,22(5):869-875.
[50]廖 敏,張海军,谢晓梅.拟除虫菊酯类农药残留降解菌产气肠杆菌的分离、鉴定及降解特性研究[J]. 环境科学,2009,30(8):2445-2451.
[51]張松柏,张德咏,罗香文,等. 一株高效降解氯氰菊酯细菌的分离鉴定及降解特性[J]. 中国农学通报,2009,25(3):265-270.
[52]Hu G P,Zhao Y,Song F Q,et al. Isolation,identification and cyfluthrin-degrading potential of a novel Lysinibacillus sphaericus strain,FLQ-11-1[J]. Research in Microbiology,2014,165(2):110-118.
[53]Tallur P N,Megadi V B,Ninnekar H Z. Biodegradation of cypermethrin by Micrococcus sp. strain CPN 1[J]. Biodegradation,2008,19(1):77-82.
[54]Wang B Z,Ma Y,Zhou W Y,et al. Biodegradation of synthetic pyrethroids by Ochrobactrum tritici strain pyd-1[J]. World Journal of Microbiology & Biotechnology,2011,27(10):2315-2324.
[55]Chen S H,Hu M Y,Liu J J,et al. Biodegradation of beta-cypermethrin and 3-phenoxybenzoic acid by a novel Ochrobactrum lupini DG-S-01[J]. Journal of Hazardous Materials,2011,187(1/2/3):433-440.
[56]Saikia N,Das S K,Patel B K,et al. Biodegradation of beta-cyfluthrin by Pseudomonas stutzeri strain S1[J]. Biodegradation,2005,16(6):581-589.
[57]Zhang C,Wang S H,Yan Y C. Isomerization and biodegradation of beta-cypermethrin by Pseudomonas aeruginosa CH7 with biosurfactant production[J]. Bioresource Technology,2011,102(14):7139-7146.
[58]Selvam A,Thatheyus J,Vidhya R. Biodegradation of the synthetic pyrethroid,fenvalerate by Pseudomonas viridiflava[J]. American Journal of Microbiological Research,2013,1(2):32-38.
[59]Song H H,Zhou Z R,Liu Y X,et al. Kinetics and mechanism of fenpropathrin biodegradation by a newly isolated Pseudomonas aeruginosa sp. strain JQ-41[J]. Current Microbiology,2015,71(3):326-332.
[60]Zhang X Q,Hao X X,Huo S S,et al. Isolation and identification of the Raoultella ornithinolytica-ZK4 degrading pyrethroid pesticides within soil sediment from an abandoned pesticide plant[J]. Archives of Microbiology,2019,201(9):1207-1217.
[61]Zhang C,Jia L,Wang S H,et al. Biodegradation of beta-cypermethrin by two Serratia spp. with different cell surface hydrophobicity[J]. Bioresource Technology,2010,101(10):3423-3429.
[62]Cycoń M,Z·mijowska A,Piotrowska-Seget Z. Enhancement of deltamethrin degradation by soil bioaugmentation with two different strains of Serratia marcescens[J]. International Journal of Environmental Science and Technology,2014,11(5):1305-1316.
[63]Prashar P. Isolation and identification of cypermethrin degrading Serratia nematodiphila from cauliflower rhizosphere[J]. International Journal of Pharmtech Research,2015,7(1):217-230.
[64]Guo P,Wang B Z,Hang B J,et al. Pyrethroid-degrading Sphingobium sp. JZ-2 and the purification and characterization of a novel pyrethroid hydrolase[J]. International Biodeterioration & Biodegradation,2009,63(8):1107-1112.
[65]崔志峰,汪 华,王渭霞,等. 氯氰菊酯降解菌CY22-7的分离鉴定及降解特性研究[J]. 环境污染与防治,2009,31(11):35-38,43.
[66]Chen S H,Yang L,Hu M Y,et al. Biodegradation of fenvalerate and 3-phenoxybenzoic acid by a novel Stenotrophomonas sp. strain ZS-S-01 and its use in bioremediation of contaminated soils[J]. Applied Microbiology and Biotechnology,2011,90(2):755-767.
[67]Saikia N,Gopal M. Biodegradation of beta-cyfluthrin by fungi[J]. Journal of Agricultural and Food Chemistry,2004,52(5):1220-1223.
[68]秦 坤,朱鲁生,王金花.氯氰菊酯降解真菌的筛选及其降解特性研究[J]. 环境工程学报,2010,4(4):950-954.
[69]Chen S H,Luo J J,Hu M Y,et al. Microbial detoxification of bifenthrin by a novel yeast and its potential for contaminated soils treatment[J]. PLoS One,2012,7(2):e30862.
[70]Chen S H,Hu Q B,Hu M Y,et al. Isolation and characterization of a fungus able to degrade pyrethroids and 3-phenoxybenzaldehyde[J]. Bioresource Technology,2011,102(17):8110-8116.
[71]Palmer-Brown W,de Melo S P,Murphy C D. Cyhalothrin biodegradation in Cunninghamella elegans[J]. Environmental Science and Pollution Research International,2019,26(2):1414-1421.
[72]張久刚,闫艳春. 一株氯氰菊酯降解菌16S rDNA,gyrB和GyrB的系统发育分析[J]. 生物信息学,2008,6(2):55-58.
[73]张玲玲,崔德杰,洪永聪,等. 氯氰菊酯降解放线菌的分离与筛选[J]. 青岛农业大学学报(自然科学版),2008,25(4):280-284.
[74]Lin Q S,Chen S H,Hu M Y,et al. Biodegradation of cypermethrin by a newly isolated actinomycetes HU-S-01 from wastewater sludge[J]. International Journal of Environmental Science and Technology,2011,8(1):45-56.
[75]Chen S H,Lai K P,Li Y A,et al. Biodegradation of deltamethrin and its hydrolysis product 3-phenoxybenzaldehyde by a newly isolated Streptomyces aureus strain HP-S-01[J]. Applied Microbiology and Biotechnology,2011,90(4):1471-1483.
[76]Chen S H,Geng P,Xiao Y,et al. Bioremediation of β-cypermethrin and 3-phenoxybenzaldehyde contaminated soils using Streptomyces aureus HP-S-01[J]. Applied Microbiology and Biotechnology,2012,94(2):505-515.
[77]Arbeli Z,Fuentes C L. Accelerated biodegradation of pesticides:an overview of the phenomenon,its basis and possible solutions and a discussion on the tropical dimension[J]. Crop Protection,2007,26(12):1733-1746.
[78]Maloney S E,Maule A,Smith A R. Purification and preliminary characterization of permethrinase from a pyrethroid-transforming strain of Bacillus cereus[J]. Applied and Environmental Microbiology,1993,59(7):2007-2013.
[79]Liang W Q,Wang Z Y,Li H,et al. Purification and characterization of a novel pyrethroid hydrolase from Aspergillus niger ZD11[J]. Journal of Agricultural and Food Chemistry,2005,53(19):7415-7420.
[80]Wu P C,Liu Y H,Wang Z Y,et al. Molecular cloning,purification,and biochemical characterization of a novel pyrethroid-hydrolyzing esterase from Klebsiella sp. strain ZD112[J]. Journal of Agricultural and Food Chemistry,2006,54(3):836-842.
[81]Tang A X,Liu H,Liu Y Y,et al. Purification and characterization of a novel β-Cypermethrin-degrading aminopeptidase from Pseudomonas aeruginosa GF31[J]. Journal of Agricultural and Food Chemistry,2017,65(43):9412-9418.
[82]Rajavat A S,Rai S,Pandiyan K,et al. Sustainable use of the spent mushroom substrate of Pleurotus Florida for production of lignocellulolytic enzymes[J]. Journal of Basic Microbiology,2020,60(2):173-184.
[83]Li G,Wang K,Liu Y H. Molecular cloning and characterization of a novel pyrethroid-hydrolyzing esterase originating from the Metagenome[J]. Microbial Cell Factories,2008,7(1):38.
[84]Wang B Z,Guo P,Hang B J,et al. Cloning of a novel Pyrethroid-Hydrolyzing carboxylesterase gene from Sphingobium sp. strain JZ-1 and characterization of the gene product[J]. Applied and Environmental Microbiology,2009,75(17):5496-5500.
[85]Zhai Y,Li K,Song J L,et al. Molecular cloning,purification and biochemical characterization of a novel pyrethroid-hydrolyzing carboxylesterase gene from Ochrobactrum anthropi YZ-1[J]. Journal of Hazardous Materials,2012,221-222(4):206-212.
[86]Fan X J,Liu X L,Huang R,et al. Identification and characterization of a novel thermostable pyrethroid-hydrolyzing enzyme isolated through metagenomic approach[J]. Microbial Cell Factories,2012,11(1):33.
[87]Ruan Z Y,Zhai Y,Song J L,et al. Molecular cloning and characterization of a newly isolated pyrethroid-degrading esterase gene from a genomic library of Ochrobactrum anthropi YZ-1[J]. PLOS One,2013,8(10):e77329.
[88]Chen S H,Lin Q S,Xiao Y,et al. Monooxygenase,a novel beta-cypermethrin degrading enzyme from Streptomyces sp.[J]. PLoS One,2013,8(9):e75450.
[89]Wei T,Feng S X,Shen Y L,et al. Characterization of a novel thermophilic pyrethroid-hydrolyzing carboxylesterase from Sulfolobus tokodaii into a new family[J]. Journal of Molecular Catalysis B-Enzymatic,2013,97(1):225-232.
[90]Cai X H,Wang W,Lin L,et al. Autotransporter domain-dependent enzymatic analysis of a novel extremely thermostable carboxylesterase with high biodegradability towards pyrethroid pesticides[J]. Scientific Reports,2017,7(1):3461.
[91]Fan X J,Liang W Q,Li Y F,et al. Identification and immobilization of a novel cold-adapted esterase,and its potential for bioremediation of pyrethroid-contaminated vegetables[J]. Microbial Cell Factories,2017,16(1):149.
[92]Luo X W,Zhang D Y,Zhou X G,et al. Cloning and characterization of a pyrethroid pesticide decomposing esterase gene,Est3385,from Rhodopseudomonas palustris PSB-S[J]. Scientific Reports,2018,8(1):7384.
[93]Hong Y F,Jin Z,Qing H,et al. Characterization of a fenpropathrin-degreding strain and construction of a genetically engineered microorganism for simultaneous degradation of methyl parathion and fenpropathrin[J]. Journal of Environmental Management,2010,91(11):2295-2300.
[94]Stratton G W,Corke C T. Toxicity of the insecticide permethrin and some degradation products towards algae and cyanobacteria[J]. Environmental Pollution,1982,29(1):71-80.
[95]Zhao J Y,Chen X F,Jia D Y,et al. Identification of fungal enzymes involving 3-phenoxybenzoic acid degradation by using enzymes inhibitors and inducers[J]. MethodsX,2020,7(1):100772.
[96]Halden R U,Tepp S M,Halden B G,et al. Degradation of 3-phenoxybenzoic acid in soil by Pseudomonas pseudoalcaligenes POB310(pPOB) and two modified Pseudomonas strains[J]. Applied and Environmental Microbiology,1999,65(8):3354-3359.
[97]Dewailly E,Forde M,Robertson L,et al. Evaluation of pyrethroid exposures in pregnant women from 10 Caribbean countries[J]. Environment International,2014,63:201-206.
[98]Sun H,Chen W,Xu X L,et al. Pyrethroid and their metabolite,3-phenoxybenzoic acid showed similar (anti)estrogenic activity in human and rat estrogen receptor α-mediated reporter gene assays[J]. Environmental Toxicology and Pharmacology,2014,37(1):371-377.
[99]Tang J,Liu B,Shi Y,et al. Isolation,identification,and fenvalerate-degrading potential of Bacillus licheniformis CY-012[J]. Biotechnology & Biotechnological Equipment,2018,32(3):574-582.
[100]Zhu Y T,Li J L,Yao K,et al. Degradation of 3-phenoxybenzoic acid by a filamentous fungus Aspergillus oryzae M-4 strain with self-protection transformation[J]. Applied Microbiology and Biotechnology,2016,100(22):9773-9786.
[101]Engesser K H,Fietz W,Fischer P,et al. Dioxygenolytic cleavage of aryl ether bonds:1,2-dihydro-1,2-dihydroxy-4-carboxybenzophenone as evidence for initial 1,2-dioxygenation in 3-and 4-carboxy biphenyl ether degradation[J]. FEMS Microbiology Letters,1990,57(3):317-321.
[102]Zhang J,Lang Z F,Zheng J W,et al. Sphingobium jiangsuense sp. nov.,a 3-phenoxybenzoic acid-degrading bacterium isolated from a wastewater treatment system[J]. International Journal of Systematic and Evolutionary Microbiology,2012,62(4):800-805.
[103]袁懷瑜.黑曲霉YAT1降解氯氰菊酯及3-苯氧基苯甲酸特性和途径的初步研究[D]. 雅安:四川农业大学,2012:1-14.
[104]Deng W Q,Lin D R,Yao K,et al. Characterization of a novel β-cypermethrin-degrading Aspergillus niger YAT strain and the biochemical degradation pathway of β-cypermethrin[J]. Applied Microbiology and Biotechnology,2015,99(19):8187-8198.
[105]徐 冉,魏 宁,黄 虹,等. 天然除虫菊酯与拟除虫菊酯的对比及发展建议[J]. 环境污染与防治,2019,41(9):1114-1119.
