大连两个海区海带相关可培养细菌群落分析
2017-12-18孙丕海杨志平李晓丽王连顺孙雪莹马悦欣
孙丕海,钱 坤,杨志平,李晓丽,王连顺,孙雪莹,马悦欣
( 1.大连海洋大学,海珍品苗种培育基地,辽宁 大连116023;2.大连金普新区海洋与渔业局,辽宁 大连116100;3.大连汇新钛设备开发有限公司,辽宁 大连116039;4.大连海洋大学,农业部北方海水增养殖重点实验室,辽宁 大连116023 )
大连两个海区海带相关可培养细菌群落分析
孙丕海1,钱 坤2,杨志平3,李晓丽4,王连顺4,孙雪莹4,马悦欣4
( 1.大连海洋大学,海珍品苗种培育基地,辽宁 大连116023;2.大连金普新区海洋与渔业局,辽宁 大连116100;3.大连汇新钛设备开发有限公司,辽宁 大连116039;4.大连海洋大学,农业部北方海水增养殖重点实验室,辽宁 大连116023 )
为研究大连两个海区海带相关可培养细菌群落,于2014年12月和2015年3月分别自登沙河湾和正明寺海带养殖区采集健康海带孢子体和海水,采用2216E涂布平板法,共分离出240株细菌,对所有菌株进行PCR和16S rDNA测序。结果表明,120条海带相关序列归属为4个门、5个纲、8个目、15个科、19个属、42个种;120条海水序列属于4个门、6个纲、10个目、18个科、26个属,46个种。在门水平,海带相关和海水细菌群落均可分为变形菌门、厚壁菌门、放线菌门和拟杆菌门,其中变形菌门和厚壁菌门为优势门。在属水平,海带相关优势属为嗜冷杆菌属、假单胞菌属、副球菌属和Salinibacterium;海水相关优势属为芽孢杆菌属、假单胞菌属、动球菌属、假交替单胞菌属和弧菌属。只从海带分离出的细菌属于微杆菌属、微小杆菌属、黄杆菌属、短小杆菌属、肠杆菌属、寡养单胞菌属和原小单孢菌属。Sørensen指数表明,两个海区海带细菌群落有一定差异,不同海区海水及同一海区海带与海水细菌群落明显不同。
海带孢子体;16S rDNA;细菌群落
海带(Saccharinajaponica,原名Laminariajaponica)[1]是一种经济价值较高的大型褐藻, 广泛应用于食品、医药、化工等方面,是大连地区大规模栽培的主要藻类之一。Duan等[2]对胶州湾海带表面进行了扫描电子显微镜观察和细菌的分离鉴定。从团岛湾潮间带采集的海带表面分离的细菌种类比较简单[3]。Balakirev等[4]分析发现太平洋西北地区广泛分布的海带典型种及其常见形态变种共生微生物群落存在显著差异,认为高遗传相似性形式(海带典型种和常见形态变种)的表型差异由宿主—共生体相互作用引起,细菌共生体可以作为区分遗传相似藻类形式的敏感标记。而假交替单胞菌属(Pseudoalteromonas)、弧菌属(Vibrio)和盐单胞菌属(Halomonas)与海带病害相关[5-8]。不过目前有关海带相关细菌群落的研究仍然较少。本试验采用传统培养方法对大连登沙河湾和正明寺养殖区健康海带相关和海水细菌群落进行了比较研究,以期为了解细菌与海带的相互作用提供参考。
1 材料与方法
1.1 样品采集
分别于2014年12月和2015年3月自大连登沙河湾和正明寺养殖区采集健康海带孢子体(1棵)和海水(3个点混合样)。
1.2 细菌分离纯化
剪取海带叶片生长点附近叶片10 g,经无菌海水冲洗3遍后,按一定比例加入无菌海水磨碎后作为原液,梯度稀释后涂布2216E平板;水样直接作为原液,梯度稀释后涂布2216E平板;室温(约15 ℃)培养14 d,从各样品适当稀释度平板上各随机挑取30个菌落,在2216E平板上分离纯化,获得纯培养于2216E斜面4 ℃保存。
1.3 细菌16S rRNA基因测序
将待测菌株接种2216E液体培养基过夜培养,使用生工生物工程(上海) 股份有限公司的土壤基因组DNA快速提取试剂盒提取细菌基因组DNA,提取方法见试剂盒说明书。用引物为F27/R1492[9]扩增细菌16S rRNA基因片段,用1% (m/V)琼脂糖凝胶电泳检测PCR产物,将PCR 产物送生工生物工程(上海) 股份有限公司测序。
1.4 细菌16S rRNA基因序列分析
将各菌株序列于EzBioCloud(http://www.ezbiocloud.net/eztaxon)提供的公共数据库中进行序列同源性比对,从结果中取相似性最高的序列。将比对后相似性大于97%的序列作为1个种[10],反之则为2个。
为了比较两个海区海带和海水细菌群落,计算Sørensen相似性指数[11]。
2 结果与分析
2.1 海带和海水可培养细菌群落组成分析
自海带及其周围海水各分离可培养细菌120株。经过16S rRNA基因序列比对,120条海带相关序列归属为4个门(图1)、5个纲(图2)、8个目、15个科(图3)、19个属(图4);120条海水序列属于4个门(图1)、6个纲(图2)、10个目、18个科(图3)、26个属(图4)。
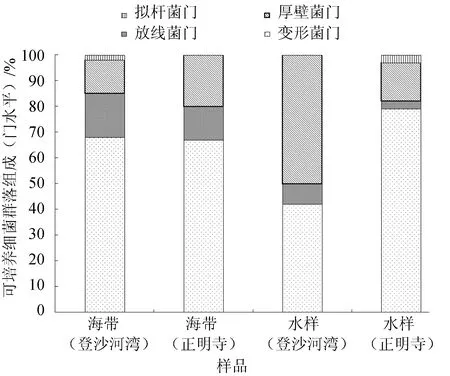
图1 海带和海水可培养细菌群落组成(门水平)
由图1可见,登沙河湾海带优势门为变形菌门(68.33%)、放线菌门(16.67%)和厚壁菌门(13.33%);正明寺海带优势门为变形菌门(66.67%)、厚壁菌门(20.00%)和放线菌门(13.33%);登沙河湾海区水样优势门为厚壁菌门(50.00%)和变形菌门(41.67%);正明寺海区水样优势门为变形菌门(78.33%)和厚壁菌门(15.00%)。
由图2可见,登沙河湾海带优势纲为γ-变形菌纲(56.67%)、放线菌纲(16.67%)、芽孢杆菌纲(13.33%)和α-变形菌纲(11.67%);正明寺海带优势纲为γ-变形菌纲(45.00%)、α-变形菌纲(21.67%)、芽孢杆菌纲(20.00%)和放线菌纲(13.33%);登沙河湾海区水样优势纲依次为芽孢杆菌纲(51.67%)和γ-变形菌纲(31.67%);正明寺海区水样优势纲依次为γ-变形菌纲(68.33%)、芽孢杆菌纲(15.00%)和α-变形菌纲(10.00%)。
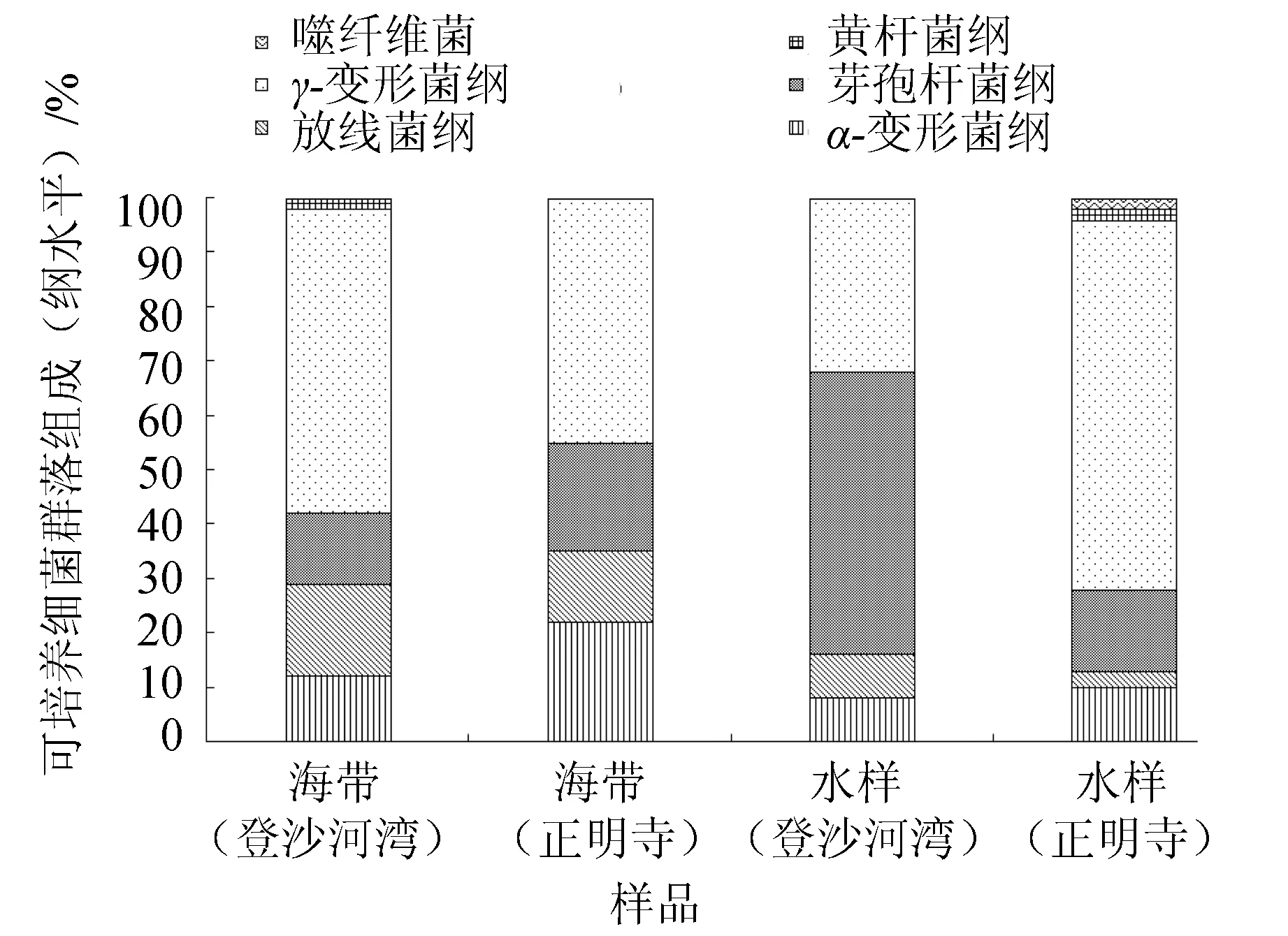
图2 海带和海水可培养细菌群落组成(纲水平)
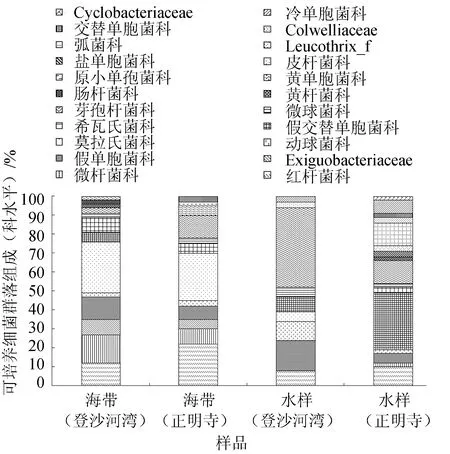
图3 海带和海水可培养细菌群落组成(科水平)
由图3可见,登沙河湾海带优势科依次为莫拉氏菌科(26.67%)、微杆菌科(15.00%)、假单胞菌科(13.33%)和红杆菌科(11.67%);正明寺海带优势科为莫拉氏菌科(25.00%)、红杆菌科(21.67%)和芽孢杆菌科(11.67%);登沙河湾海区水样优势科依次为芽孢杆菌科(41.67%)、假单胞菌科(15.00%)和动球菌科(10.00%);正明寺海区水样优势科依次为假交替单胞菌科(30.00%)、芽孢杆菌科(13.33%)、弧菌科(11.67%)和红杆菌科(10.00%)。
由图4可见,登沙河湾海带优势属依次为嗜冷杆菌属(Psychrobacter)(26.67%)、假单胞菌属(Pseudomonas)(13.33%)、副球菌属(Paracoccus) (11.67%)和Salinibacterium(11.67%); 正明寺海带优势属为嗜冷杆菌属(25.00%)和副球菌属(20.00%);登沙河湾海区水样优势属依次为芽孢杆菌属(Bacillus)(35.00%)、假单胞菌属(15.00%)和动球菌属(Planococcus)(10.00%);正明寺海区水样优势属依次为假交替单胞菌属(30.0%)、芽孢杆菌属(13.33%)和弧菌属(10.00%)。
所有菌株与其相关有效发表菌株的序列相似性为99%。按16S rRNA基因序列相似性大于97%的菌株归于同一种计[10],从海带和海水分离的细菌分别可以归为42个种和46个种(表1)。
2.2 海带和海水细菌群落比较
Sørensen指数可反映不同细菌群落的相似性,Sørensen指数值越小,说明群落间相似性越低,群落间差异越大。两个海区海带和海水细菌群落之间Sørensen相似性指数结果见表2,不同海区海带细菌群落有一定差异,不同海区海水和同一海区海带与海水细菌群落明显不同。
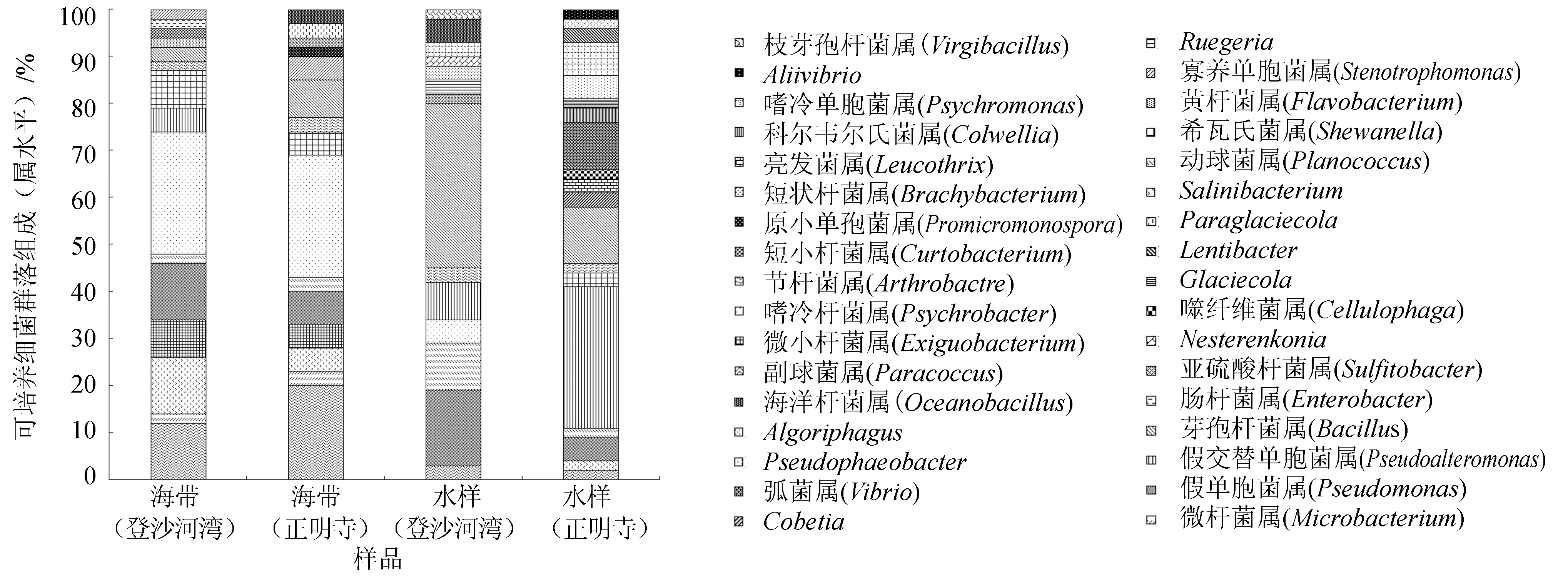
图4 海带和海水可培养细菌群落组成(属水平)

样品细菌群落组成(种水平)登沙河湾海带地下节杆菌(Arthrobactersubterraneus),Bacillusoryzaecorticis,B.tianshenii,萎蔫短小杆菌(Curtobacteriumflaccumfaciens),Enterobacterxiangfangensis,Exiguobacteriumoxidotolerans,E.sibiricum,Flavobacteriumfrigidarium,Microbacteriumfoliorum,Oceanobacilluspicturae,Paracoccusaestuarii,P.haeundaensis,马氏副球菌(P.marcusii),P.seriniphilus,Planococcusmaritimus,埃氏假交替单胞菌(Pseudoalteromonasespejiana),Pseudomonasgessardii,P.koreensis,Psychrobacteraquimaris,P.fozii,P.nivimaris,解蛋白嗜冷杆菌(P.proteolyticus),阿穆尔斯克湾盐地杆菌(Salinibacteriumamurskyense),Shewanellaarctica,S.glacialipiscicola,S.vesiculosa,嗜根寡养单胞菌(Stenotrophomonasrhizophila)正明寺海带贝热尔氏节杆菌(Arthrobacterbergerei),海口芽孢杆菌(Bacillushaikouensis),B.oryzaecorticis,Exiguobacteri-umoxidotolerans,E.sibiricu,栖稻黄色单胞菌(Flavimonasoryzihabitans),Microbacteriummaritypicum,氧化微杆菌(M.oxydans),Oceanobacilluspicturae,Paracoccusaestuarii;P.homiensis,P.seriniphilus,Paragla-ciecolamesophila,Planococcusmaritimus,P.plakortidis,Promicromonosporaxylanilytica,Pseudomonasgra-nadensis,Psychrobacteraquimaris,P.arcticus,P.cryohalolentis,P.nivimaris,P.okhotskensis,P.proteolyti-cus,嗜根寡养单胞菌(Salinibacteriumamurskyense),Shewanellaarctica,S.basaltis,S.vesiculosa,嗜根寡养单胞菌(Stenotrophomonasrhizophila),Sulfitobacterlitoralis登沙河湾海区水样Arthrobacterarilaitensis,海口芽孢杆菌(Bacillushaikouensis),B.oceanisediminis,B.oryzaecorticis,B.vietna-mensis,Brachybacteriumginsengisoli,Nesterenkoniajeotgali,马氏副球菌(Paracoccusmarcusii),P.seriniphi-lus,Planococcusrifietoensis,Planomicrobiumchinense,P.koreense,P.okeanokoites,北极假交替单胞菌(Pseud-oalteromonasarctica),产黑假交替单胞菌(P.nigrifaciens),Pseudomonascoleopterorum,P.rhodesiae,猴假单胞菌(P.simiae),Psychrobactercibarius,P.nivimaris,肺嗜冷杆菌(P.pulmonis),Ruegeriaatlantica,Sulfito-bacterguttiformis,Virgibacillusnecropolis正明寺海区水样Algoriphagusaquimarinus,Aliivibriologei,Arthrobacterhumicola,海口芽孢杆菌(Bacillushaikouensis),B.litoralis,B.oryzaecorticis,B.vietnamensis,Cellulophagabaltica,Cobetialitoralis,Colwelliaasteriadis,C.meonggei,Glaciecolapunicea,浒苔缓生菌(Lentibacteralgarum),毛霉亮发菌(Leucothrixmucor),Paracoccuszeaxanthinifaciens,Planococcusdechangensis,北极假交替单胞菌(Pseudoalteromonasarctica),埃氏假交替单胞菌(P.espejiana),产黑假交替单胞菌(P.nigrifaciens),Pseudomonasrhodesiae,猴假单胞菌(P.simiae),Pseu-dophaeobacterarcticus,Psychromonasarctica,阿穆尔斯克湾盐地杆菌(Salinibacteriumamurskyense),海动物肠希瓦氏菌(Shewanellamarinintestina),Vibriohemicentroti,V.lentus,灿烂弧菌(V.splendidus)
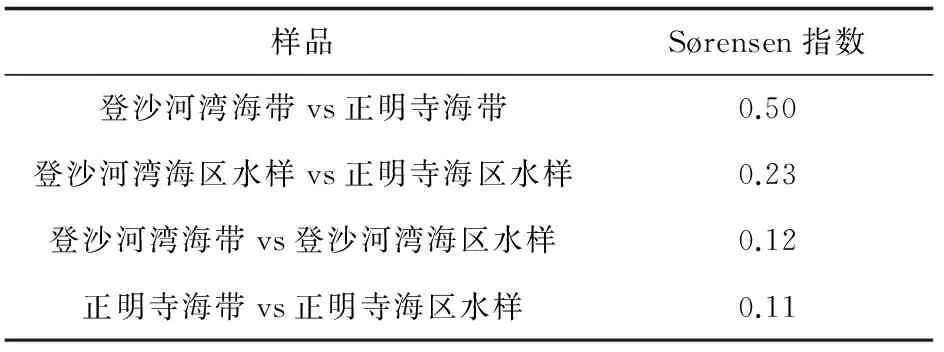
表2 海带和海水可培养细菌群落的Sørensen指数
3 讨 论
3.1 海带相关可培养细菌群落组成分析
已有研究表明,海带相关细菌群落包括变形菌门、拟杆菌门(CFB 群)、放线菌门和厚壁菌门[2-4,12]。在门水平,本试验从海带分离的细菌和前人研究结果基本一致。海带属的其他种如L.saccharina相关细菌群落也属于上述四个门[13-14],L.digitata附生细菌群落由变形菌门、放线菌门和拟杆菌门组成[15],L.longicruris中发现变形菌门和拟杆菌门[16],细布海带(L.religiosa)中只有变形菌门[17],而L.hyperborea相关细菌群落也包括变形菌门、拟杆菌门、厚壁菌门、放线菌门[18-21]。在科水平,从健康海带分离出22个科的细菌(图3),其中假单胞菌科、假交替单胞菌科、交替单胞菌科、微球菌科、莫拉氏菌科、肠杆菌科、芽孢杆菌科和黄杆菌科为已报道海带相关细菌群落成员[2,4,12,22-23]。此外,还分离出属于希瓦氏菌科、微杆菌科、黄单胞菌科、红杆菌科、动球菌科、原小单胞菌科的细菌。L.saccharina相关细菌群落由希瓦氏菌科、微杆菌科、微球菌科、红杆菌科和黄单胞菌科的成员组成[14]。从L.digitata上发现属于微杆菌科、微球菌科和红杆菌科的细菌[15]。红杆菌科也可从L.hyperborea上分离到[20]。动球菌科和原小单胞菌科作为海带属藻类相关细菌群落组成为国内外首次报道。本试验从健康海带分离出共计19个属的细菌(图4),其中芽孢杆菌属、黄杆菌属(Flavobacterium)、假交替单胞菌属、假单胞菌属和嗜冷杆菌属为已报道海带相关细菌群落成员[2,4,12]。希瓦氏菌属(Shewanella)、亚硫酸杆菌属(Sulfitobacter)、假交替单胞菌属、假单胞菌属、寡养单胞菌属(Stenotrophomonas)、Glaciecola和节杆菌属(Arthrobacter)在L.saccharina中发现[14]。L.digitata的细菌群落由短小杆菌属(Curtobacterium)、节杆菌属、副球菌属、假交替单胞菌属和假单胞菌属组成[15]。从L.longicruris分离出属于假单胞菌属和黄杆菌属的种类[16]。芽孢杆菌属的细菌也可从L.hyperborea分离获得[20]。微杆菌属在澳洲石莼(Ulvaaustralis)和Pachymeniopsislauceolata中发现[24-25]。肠杆菌属(Enterobacter)、动性球菌属和Salinibacterium分别属于藻类Cystoseiramyrica[26]、Gracilariadura[27]、Camphylaephorahyphaeoides[28]和Ahnfeltiasp. (Genbank未发表数据)相关细菌群落成员。原小单胞菌属(Promicromonospora)、Virgibacillus和Oceanobacillus作为海藻相关细菌群落成员为首次发现,扩展了目前海带相关微生物群落的知识。
海带相关细菌可能是潜在的抗菌资源,如一种芽孢杆菌、一种假单胞菌和一种黄杆菌(Flavobacteriumsp.)具有抗微生物活力[29],也可能是潜在病原,如Pseudoalteromonasbacteriolytica[6]和P.elyakovii[7]分别是海带红点病和叶斑点伤病害的病原菌。7株假交替单胞菌属和4株弧菌属细菌可能为海带孔烂病的潜在病原[8]。本研究从海带中发现了芽孢杆菌、假单胞菌、黄杆菌、假交替单胞菌和弧菌属,其生态作用及其对宿主的影响待于进一步研究。
3.2 海带和海水细菌群落比较
本试验中海带养殖区海水中的细菌群落由变形菌门、厚壁菌门、放线菌门和拟杆菌门组成。这些门广泛分布在其他海藻生长海域水体中,如L.saccharina生长海域水体中的细菌属于变形菌门和拟杆菌门[13]。澳洲石莼生长海域水体中优势门为变形菌门、拟杆菌门和放线菌门,而厚壁菌门丰度较低。尽管在门水平上海带相关细菌群落与其周围海水的细菌群落大致相似,但在种水平,海带和海水细菌群落明显不同。这一结果与已有的研究结果基本一致[13,19,30]。Staufenberger 等[13]研究结果表明,L.saccharina和海水样品DGGE 条带图谱明显不同,相似性只有20%。L.hyperborea表面生物膜和其周围海水细菌群落重叠较少[19]。Burke等[30]通过16S rRNA基因克隆文库技术发现在门水平澳洲石莼附生细菌群落和和其周围海水细菌群落类似,但是,Bray-Curtis相似性和聚类分析结果表明,在较低等分类水平(种),两细菌群落明显不同。可能是由于两种环境的物理差异引起群落组成的差异。海藻上细菌的可能来源之一是其周围的海水,因此海藻表面细菌可能是通过选择过程或经直接表面接触输送细菌,从海水较少的种群中吸收成员[19]。
[1] Lane C E, Mayes C, Druehl L D, et al. A multi-gene molecular investigation of the kelp (Laminariales, Phaeophyceae) supports substantial reorganization [J]. Journal of Phycology, 2006, 42(2):493-512.
[2] Duan D, Xu L, Fei X, et al. Marine organisms attached to seaweed surfaces in Jiaozhou Bay, China[J]. World Journal of Microbiology and Biotechnology, 1995, 11(3):351-352.
[3] Wang Z F, Xiao T, Pang S J, et al. Isolation and identification of bacteria associated with the surfaces of several algal species [J]. Chinese Journal of Oceanology and Limnology, 2009, 27(3):487-492.
[4] Balakirev E S, Krupnova T N, Ayala F J. Symbiotic associations in the phenotypically-diverse brown algaSaccharinajaponica[J]. PloS One, 2012, 7(6):e39587.
[5] Yumoto I, Yamaguchi K, Yamada K, et al. Relationship between bacterial flora and occurrence of theAlteromonassp., the causative agent of red-spots on the culture bed of makonbuLaminariajaponica, in the coastal area of Funka Bay [J]. Nippon Suisan Gakkaishi, 1989, 55(11):1907-1914.
[6] Sawabe T, Makino H, Tatsumi M, et al.Pseudoalteromonasbacteriolyticasp. nov., a marine bacterium that is the causative agent of red spot disease ofLaminariajaponica[J]. International Journal of Systematic Bacteriology, 1998, 48(3):769-774.
[7] Sawabe T, Tanaka R, Christen R. et al. Assignment ofAlteromonaselyakoviiKMM162T and five strains isolated from spot wounded fronds ofLaminariajaponicatoPseudoalteromonaselyakoviicomb. nov. and the extended description of the species [J]. International Journal of Systematic and Evolutionary Microbiology, 2000, 50(1):265-271.
[8] Wang G, Shuai L, Li Y, et al. Phylogenetic analysis of epiphytic marine bacteria on Hole-Rotten diseased sporophytes ofLaminariajaponica[J]. Journal of Applied Phycology, 2008, 20(4):403-409.
[9] Stackebrandt E, Goodfellow M. Nucleic Acid Techniques in Bacterial Systematics [M]. Chichester:John Wiley & Sons Press,1991:115-175.
[10] Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology [J]. International Journal of Systematic Bacteriology, 1994, 44(4):846-849.
[11] Sørensen T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons [J]. Kongelige Danske Videnskabernes Selskabs Biologiske Skrifter, 1948, 5(4):1-34.
[12] Dimitrieva G Y, Crawford R L, Yuksel G Ü. The nature of plant growth-promoting effects of a pseudoalteromonad associated with the marine algaLaminariajaponicaand linked to catalase excretion [J]. Journal of Applied Microbiology, 2006, 100(5):1159-1169.
[13] Staufenberger T, Thiel V, Wiese J, et al. Phylogenetic analysis of bacteria associated withLaminariasaccharina[J]. FEMS Microbiology Ecology, 2008, 64(1):65-77.
[14] Wiese J,Thiel V, Nagel K, et al. Diversity of antibiotic-active bacteria associated with the brown algaLaminariasaccharinafrom the Baltic Sea [J]. Marine Biotechnology, 2009, 11(2):287-300.
[15] Salaün S, Nelly K, Philippe P, et al. Whole-cell spectroscopy is a convenient tool to assist molecular identification of cultivatable marine bacteria and to investigate their adaptive metabolism [J]. Talanta, 2010, 80(5):1758-1770.
[16] Laycock R A. The detrital food chain based on seaweeds. I. Bacteria associated with the surface ofLaminariafronds [J]. Marine Biology, 1974, 25(3):223-231.
[17] Vairappan C S, Suzuki M, Motomura T, et al. Pathogenic bacteria associated with lesions and thallus bleaching symptoms in the Japanese kelpLaminariareligiosaMiyabe (Laminariales, Phaeophyceae) [J]. Hydrobiologia, 2001, 445(1):183-191.
[18] Bengtsson M M, Øvreås L. Planctomycetes dominate biofilms on surfaces of the kelpLaminariahyperborea[J]. BMC Microbiology, 2010, 10(3):1-12.
[19] Bengtsson M M, Sjotun K, Ovreas L, et al. Seasonal dynamics of bacterial biofilms on the kelpLaminariahyperborea[J]. Aquatic Microbial Ecology, 2010, 60(1):71-83.
[20] Bengtsson M M, Sjøtun K, Storesund J E, et al. Utilization of kelp-derived carbon sources by kelp surface-associated bacteria [J]. Aquatic Microbial Ecology, 2011, 62(2):191-199.
[21] Bengtsson M M, Sjøtun K, Lanzén A, et al. Bacterial diversity in relation to secondary production and succession on surfaces of the kelpLaminariahyperborea[J]. The ISME Journal, 2012, 6(12):2188-2198.
[22] 徐涤,秦松,庞国兴,等. 青岛和大连海区海带(Laminariajaponica) 外生细菌的16SrRNA 基因序列分析[J].高技术通讯, 2004,14(2):81-86.
[23] Hollants J, Leliaert F, De Clerck O, et al. What we can learn from sushi: a review on seaweed bacterial associations [J]. FEMS Microbiology Ecology, 2013, 83(1):1-16.
[24] Kanagasabhapathy M, Sasaki H, Nagata S. Phylogenetic identification of epibiotic bacteria possessing antimicrobial activities isolated from red algal species of Japan [J]. World Journal of Microbiology Biotechnology, 2008, 24(10):2315-2321.
[25] Burmølle M, Webb J S, Rao D, et al. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms [J]. Applied and Environmental Microbiology, 2006, 72(6):3916-3923.
[26] Mohammed N A, Hassan H M, Rateb M E, et al. Diketopiperazine derivatives fromEnterobactercloacaeisolated from the Red Sea algaCystoseiramyrica[J]. Egyptian Pharmaceutical Journal, 2013, 12(2):163.
[27] Singh R P, Bijo A J, Baghel R S, et al. Role of bacterial isolates in enhancing the bud induction in the industrially important red algaGracilariadura[J]. FEMS Microbiology Ecology, 2011, 76(2):381-392.
[28] Beleneva I A, Zhukova N V. Bacterial communities of some brown and red algae from Peter the Great Bay, the Sea of Japan [J]. Microbiology, 2006, 75(3):348-357.
[29] Zheng L, Han X, Chen H, et al. Marine bacteria associated with marine macroorganisms: the potential antimicrobial resources[J].Annals Microbiology,2005,55(2):119-124.
[30] Burke C, Thomas T, Lewis M, et al. Composition, uniqueness and variability of the epiphytic bacterial community of the green algaUlvaaustralis[J]. ISME Journal,2011, 5(4):590-600.
CulturableBacteriaCommunityAssociatedwithSaccharinajaponicafromTwoSeaAreasinDalian
SUN Pihai1, QIAN Kun2, YANG Zhiping3, LI Xiaoli4, WANG Lianshun4, SUN Xueying4, MA Yuexin4
( 1.Seafood Seedling Breeding Base, Dalian Ocean University, Dalian 116023, China;2. Dalian Jinzhou District Oceanic and Fishery Administration, Dalian 116100, China; 3. Dalian Huixin Titanium Equipment Development Company Limited, Dalian 116039, China; 4. Key Laboratory of Mariculture and Stock Enhancement in North Chinas Sea, Ministry of Agriculture, Dalian Ocean University, Dalian 116023,China )
The aim of the study was to examine the bacterial community associated with the kelpSaccharinajaponicafrom two sea areas in Dalian. In December 2014 and March 2015, a total of 240 isolates were obtained from the healthy kelp and the surrounding seawater collected from the sea areas of Dengshahe bay and Zhengmingsi using spread plate method relied on Zobell 2216E medium. Polymerase chain reaction and partial 16S rDNA sequencing of DNA from all isolates revealed that 120 kelp-associated sequences were classified as 4 phyla, 5 classes, 8 orders, 15 families, 19 genera and 42 species; that 120 water-derived sequences were classified as 4 phyla, 6 classes, 10 orders, 18 families, 26 genera and 46 species. At phylum level,S.japonicaand seawater communities were composed of Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes, with dominant phyla of Proteobacteria and Firmicutes. At genus level, the composition of community associated withS.japonicawas represented principally byPsychrobacter,Pseudomonas,SalinibacteriumandParacoccus; the seawater community was dominated byBacillus,Pseudoalteromonas,Pseudomonas,PlanococcusandVibrio. Additionally,Microbacterium,Exiguobacterium,Flavobacterium,Curtobacterium,Enterobacter,StenotrophomonasandPromicromonosporawere exclusively present in community ofS.japonica. As indicated by Sørensen index, there were some differences in bacterial communities between kelp samples from the two different sea areas; while there existed significant differences in communities between seawater samples from the two sea areas and between kelp sample and seawater sample from the same sea area.
Saccharinajaponica;16S rDNA; bacterial community
10.16378/j.cnki.1003-1111.2017.04.016
S917.1
A
1003-1111(2017)04-0498-06
2016-03-10;
2016-12-16.
大连市海洋与渔业局项目(2014115,2014118).
孙丕海(1959-),男,高级工程师;研究方向:海珍品种苗培育及养殖.E-mail:dlsph@aliyun.com.通讯作者:马悦欣(1963-),女,教授,博士;研究方向:微生物生态学. E-mail:mayuexin@dlou.edu.cn.
