Rapid detection of Shigella and Salmonella in rhesus monkeys by loop-mediated isothermal amplification assay
2016-04-14叶莉,吴芳,WANGYi-jia等
研究报告
Rapid detection ofShigellaandSalmonellain rhesus monkeys by loop-mediated isothermal amplification assay
YE Li1△, WU Fang2△, WANG Yi-jia3, ZHENG Jun-wen3, FAN Jun-wen1,DING Ming1, SHI Yan-sheng1, ZHANG Xiao-fei4*, BAI Jie-ying1*
(1.Laboratory Animal Center, Academy of Military Medical Sciences, Beijing, 100071, China.
2. the Sixth Clinical School, Capital Medical University, Beijing 100029.
3. Capital Normal University, Beijing, 100048. 4. Beijing Institute of Veterinary Drug Control, Beijing 102600)
【Abstract】ObjectiveTo establish a loop-mediated isothermal amplification (LAMP) method for detecting diarrhea pathogens (ShigellaandSalmonella) in rhesus monkeys and evaluate the application of the LAMP method for detecting bacterial diseases in non-human primate laboratory animals. Materials and Methods A total of 205 fecal samples of rhesus monkeys were detected in this LAMP assay. The specificity and sensitivity of LAMP forShigellaandSalmonellawere analyzed, and real-time polymerase chain reaction (REAL-TIME PCR) assay was employed as control. ResultsThe LAMP method established here needed only 45 min to complete the reaction at 63℃. Its detection limit was 10 pg/μL and with a high specificity. The positive rate ofShigellaandSalmonellawas 1.5% and 6.3%, respectively. Conclusions Here we have established a fast and simpleShigellaandSalmonellaLAMP detection method that has strong specificity and high sensitivity and is suitable for rapid detection of bacterial disease in macaques. The development of this rapid detection kit is underway, and it will be helpful to the diarrhea detection.
【Key words】Shigella;Salmonella; Loop-mediated isothermal amplification, LAMP; Rhesus monkey; Bacteria detection; Diarrhea
There are hundreds of thousands of monkeys kept in cages in China as laboratory animals. According to the national rules, the monkeys should be healthy to be used in medical experiments. While many monkey farms were obsessed with diarrhea, which is common in captive Rhesus monkeys. The diarrhea is caused by bacterial infections, cage replacement, seasonal alternations, and so on. Bacterial infection is the most harmful cause, withShigellaandSalmonellaas the most common diarrhea pathogens1. These bacteria adversely affect the animals’ physical health and may infect animal keepers and other staff members. The current isolation and culturing method used for the routine diagnosis of these pathogens requires considerable time and effort. As a result, symptomatic medications cannot be promptly administered, which may result in illness delay and loss of optimal treatment timing. A monkey farm may shell out millions per year for the monkey death from diarrhea. Furthermore, blind medications can easily enhance the antimicrobial resistance of pathogens and increase difficulties in treatment. Therefore, a rapid and cheap detection technique of bacteria is urgently needed.
With the development of molecular techniques, a variety of methods such as polymerase chain reaction (PCR) and immunological methods are available to identify causative pathogens. However, these approaches require expensive equipment (PCR instrument and Multiskan Ascent) and skilled technicians. Therefore, a rapid, simple, and economical detection technique is needed for diagnosing diseases in captive rhesus monkeys.
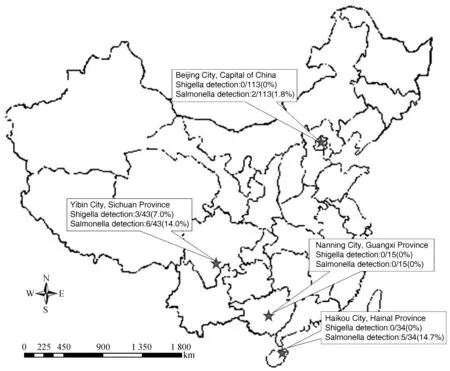
Fig. 1 Locations of the primate centers in China where macaques were investigated for Shigella and Salmonella. A total of 205 rhesus macaques feces samples were collected from Haikou, Nanning, Yibin and Beijing Cities. The results of positive rate of Shigella and Salmonella were shown in the picture.
The loop-mediated isothermal amplification (LAMP) technique is based on the strand displacement reaction and stem-loop structure that amplifies the target gene fragment under isothermal conditions.2This method requires only a water bath and the result can be judged by naked eyes. The LAMP technique can be used in rapid diagnosis of different pathogens, and some researchers have already used this technique for bacterial detection.3,4However, they were only able to obtain the LAMP reaction results using an electrophoresis map. These results are lack of real-time reaction process monitoring. Furthermore, the risk of aerosol pollution exists in the laboratory. Therefore, no accurate judgment of the process could be made to date.
In the current study, we used a Loopamp® Realtime Turbidimeter LA-320C (Eiken Chemical Co. Ltd., Tokyo, Japan) to monitor the LAMP reactions in real time and established a method that can directly detectShigellaandSalmonellain rhesus monkey feces. This method may be helpful in monitoring the healthy status of non-human primate laboratory animals.
1Materials and methods
1.1Sample collection
A total of 205 fecal samples were collected from 4 primate centers located in 4 cities (Haikou, Nanning, Yibin and Beijing) (Fig. 1). Bacterial DNA was extracted from 1 mL of cultural suspension using TIANamp Bacteria DNA Kit (Tiangen, Beijing, China). DNA was extracted from 0.1 mg feces using CHLEX reagent.
1.2Bacterial strains and cultural conditions
Standard bacteria were purchased from the National Institutes for Food and Drug Control includedSalmonellaenteritidis(CMCC 50041),Salmonellatyphi(CMCC 50071),SalmonellaTyphimurium(CMCC 50115),Shigelladysenteriae(CMCC 51252),Shigellaflexneri(CMCC 51571),Shigellaflexneri(CMCC 1.1868), andEscherichiacoli(CMCC 44102).
Our research complied with the protocols approved by the Animal Care and Use Committee of Laboratory Animal Center of the Academy of Military Medical Sciences. This research adhered to the legal requirements of China as well as the American Society of Primatologists principles for the ethical treatment of primates.
1.3Design ofShigellaandSalmonellaspecific LAMP primers
According to the nucleotide sequences ofShigellaipaHgene (M76444.1) andSalmonellainvAgene (NC_003197.1) that we retrieved from the GenBank database, LAMP primers were designed using PrimerExplorer (http://primerexplorer.jp/elamp4.0.0/index.html). Five groups of primers for these two bacteria were obtained: F3 and B3 (outer primers), FIP and BIP (inner primers), and LF or LB (loop primer).
1.4LAMP reaction condition
The LAMP reactions were performed using a Loopamp DNA Amplification Kit (Eiken Chemical Co. Ltd. Tokyo). The reaction system (25 μL) included the following: 2× reaction mix (12.5 μL), primer mix (4.6 μL), Bst DNA polymerase (1 μL), sample DNA (2 μL), distilled water (4.9 μL), and primer mix with F3, B3, FIP, BIP, and LF/LB in concentrations of 5, 5, 40, 40, and 20 pmol/μL, respectively. The reactions were performed in the Loopamp® Realtime Turbidimeter LA-320C (Eiken Chemical Co. Ltd.) for 60 min. The most optimal reaction temperature and primers were determined using a temperature gradient assay. Visual detection was performed using a fluorescent detection reagent (Eiken Chemical Co. Ltd.). The color change under normal conditions was recorded: a positive sample was green, while a negative sample was orange.
1.5LAMP assay specificity
ShigellaorSalmonellaprimers were used to detect standard bacteria (including CMCC 50041, 50071, 50115, 51252, 51571, 1.1868, and 44102). DNA was extracted as described earlier. All tests were performed in triplicate.
1.6LAMP assay sensitivity
For the sensitivity tests, DNA mixes ofShigella(CMCC51252, 51571, and 1.1868) andSalmonella(CMCC50041, 50071, 50115) were serially diluted 10-fold with sterile water from 1.0 × 10-1to 1.0 × 10-7. All tests were performed in triplicate.
1.7LAMP and real-time PCR detection
A total of 205 fecal samples from captive Rhesus monkey were examined using real-time PCR and the LAMP system which we developed for the detection ofShigellaandSalmonella. Differences were found between these two methods.
2Results
2.1Specific LAMP primers ofShigellaandSalmonella
The primer sets comprising the two outer primers (F3 and B3), two inner primers (FIP and BIP), and one loop primer (LF or LB) (Table 1) recognized six distinct regions on the ipaH ofSalmonellaand the invA ofShigellasequences. All of the primers were synthesized by Beijing AuGCT DNA-SYN Biotechnology (China).
2.2LAMP reaction temperature
The reactions were performed in the real-time turbidimeter LA-320C at 60-64℃ for 90 min. The machine detects the turbidity values every 6 s and indicates positive amplification when the turbidity values reach 0.1.5,6The earliest reaction started at 63℃, and the optimal temperature of these two bacteria was 63℃ (Fig. 2).
2.3LAMP result judgment
Fluorescent detection reagent (1 μL) was added to theShigellaandSalmonellareaction mixtures for visual detection by naked eyes. Positive amplification was green and negative amplification was orange (Fig. 3). The results also could be observed using a UV lamp (wavelength 240-260 nm or 350-370 nm), with a positive result of green and a negative result with no changes.
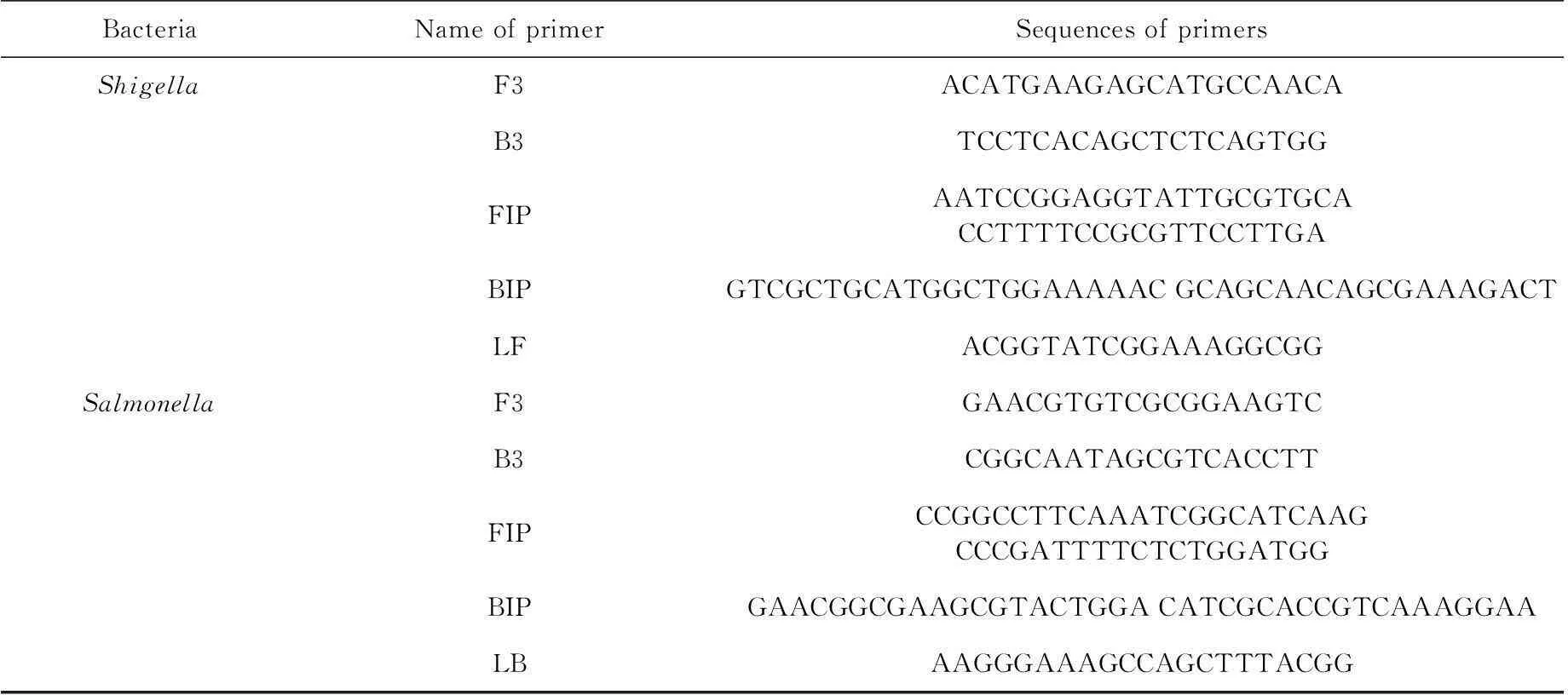
Tab. 1 The LAMP primer sequences of Shigella and Salmonella
Note. LAMP: loop-mediated isothermal amplification.
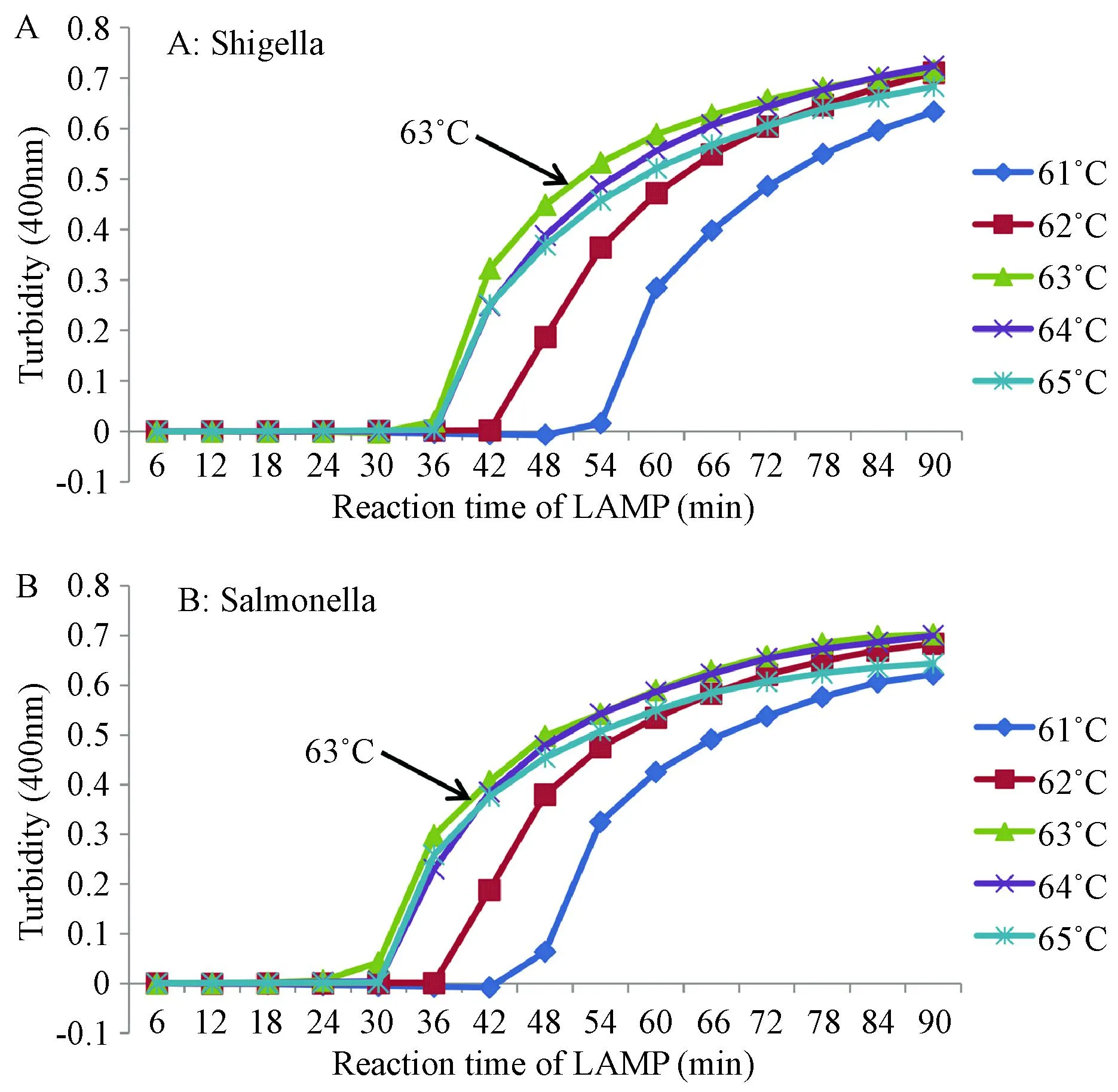
Fig. 2 The optimal temperature in detection of Shigella (A) and Salmonella (B), with LAMP reaction at 63℃.
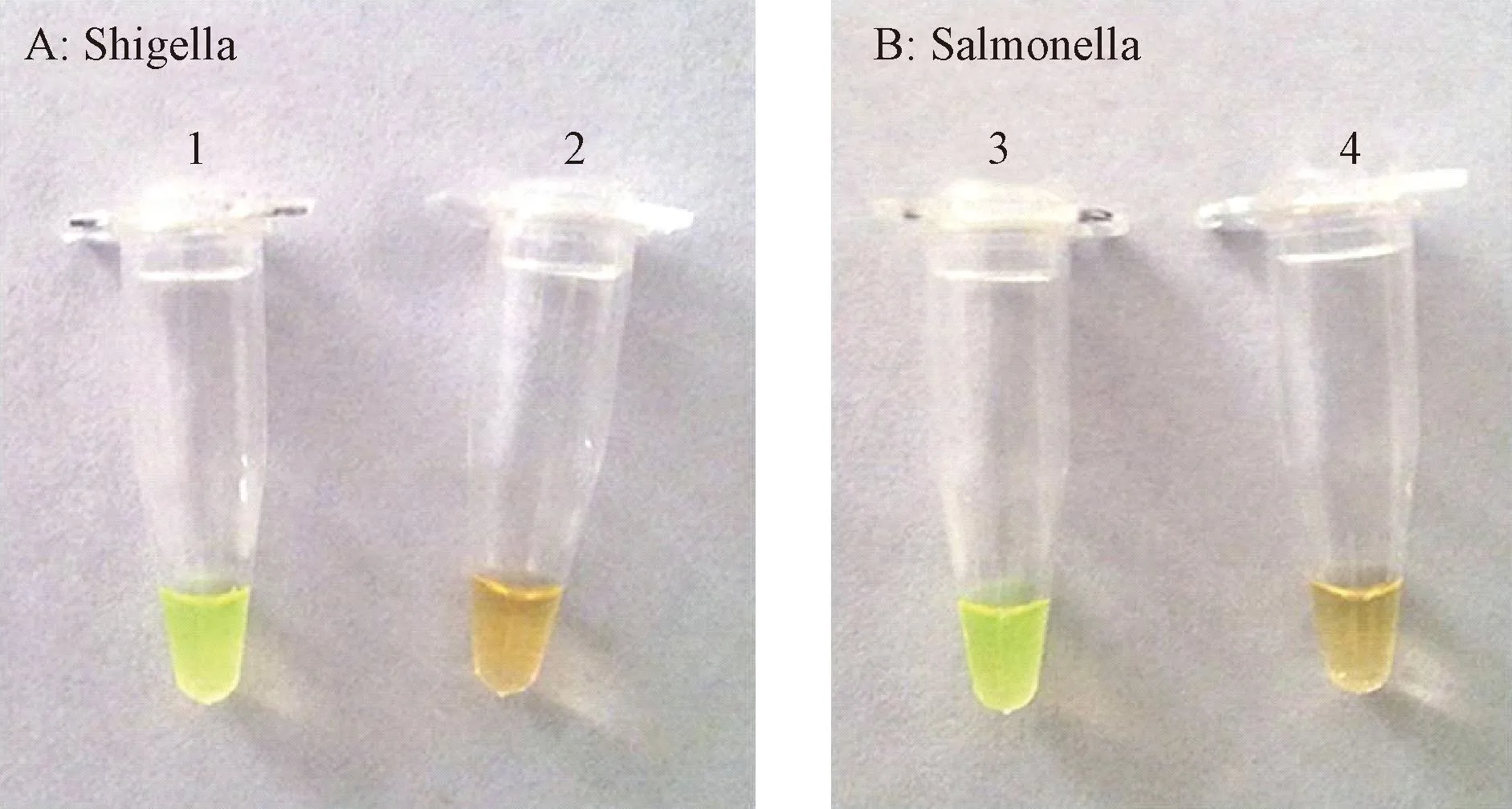
Fig. 3 Fluorescence detection of Shigella (left) and Salmonella (right) by the LAMP assay. Visual detection of Shigella (At) and Salmonella (B) in the LAMP products using fluorescence detection. 1 and 3: Positive LAMP reaction (green); 2 and 4: Negative LAMP reaction (orange).
2.4Specificity and sensitivityoftheShigellaandSalmonellaspecific LAMP assay
With theShigellaprimer, theShigellasamples were positively amplified, while theSalmonellaandE.colisamples were not (Fig. 4). TheSalmonelladetection results were the same. These results showed that the LAMP primers ofShigellaandSalmonellahad strong specificity.
ShigellaandSalmonellamixture DNA samples (212.3 ng/μL and 185.7 ng/μL, respectively) were prepared for the sensitivity assay. The loopamp® real-time turbidimeter LA-320C curve analysis showed that all positive amplifications reached a velocity curve peak >0.1 within 60 min (Fig. 5 and 6). When the DNA template concentration decreased, the reaction rate constant and response slope was decreased and the peak time delayed. The minimum detectable limits forSalmonellaandShigellawere both 10-5(DNA concentration was approximately 10 pg/μL).
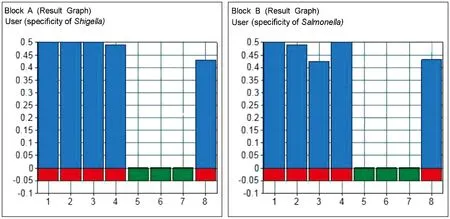
Fig. 4 Specificity of Shigella and Salmonella. With the Shigella and Salmonella primers, the samples were positively amplified, while the control samples were not. (Block A) 1, Salmonella mix (including 50041, 50071, 50115); 2, 50041; 3, 50071; 4, 50115; 5, Shigella mix (including 51252, 51571, 1.1868); 6, 44102; 7, DEPC H2O; 8, positive control. (Block B) 1, Shigella mix (including 51252, 51571, 1.1868); 2, 51252; 3, 51571; 4, 1.1868; 5, Salmonella mix (including 50041, 50071, 50115); 6, 44102; 7, DEPC H2O; 8, positive control.
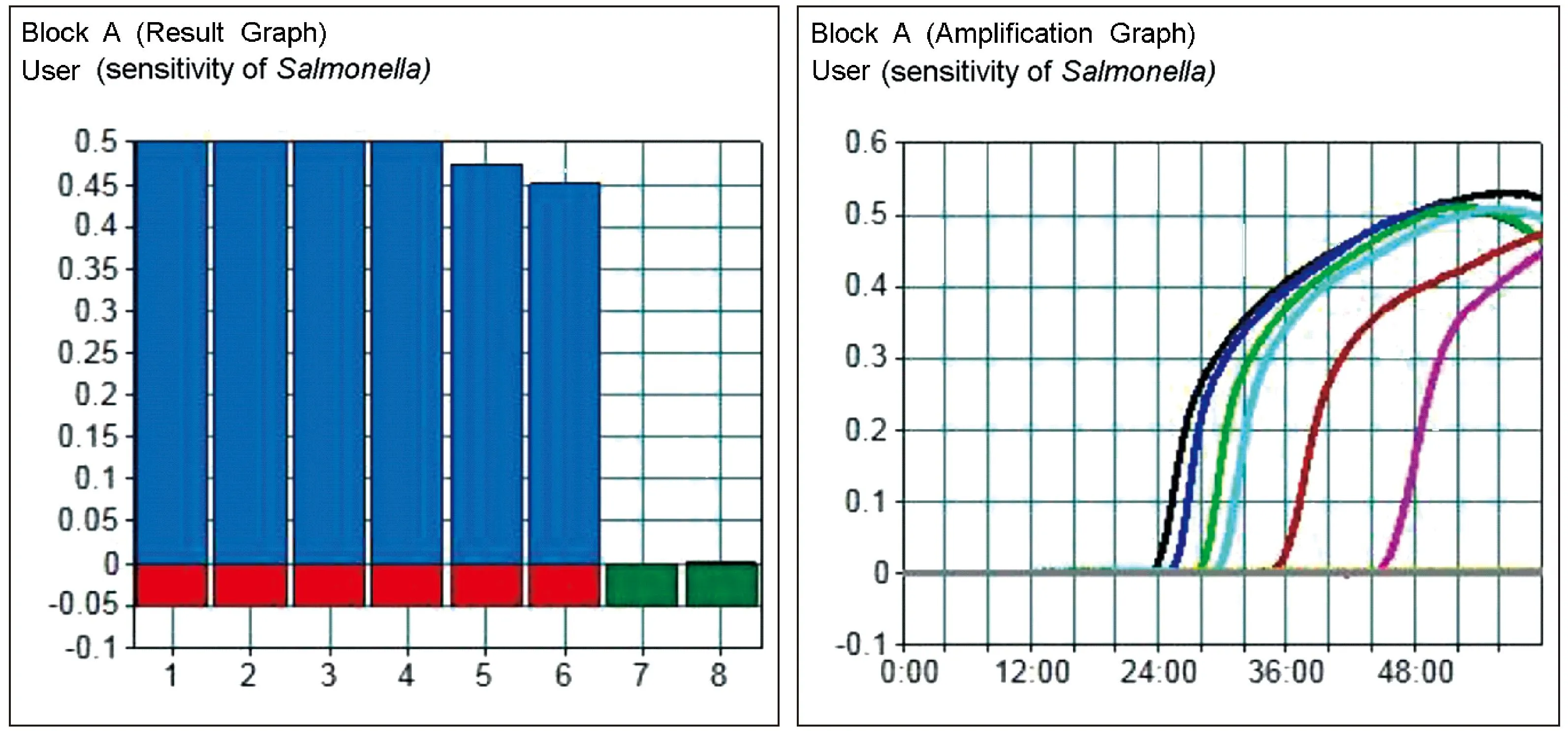
Fig. 5 Sensitivity of Salmonella in LAMP. The minimum detectable limits for Salmonella was 10-5 (DNA concentration was approximately 10 pg/μL). CH1, 1.0; CH2-CH8, 1.0 × 10-1-1.0 × 10-7.
2.5Comparison of the LAMP and real-time PCR assay
In the 205 clinical fecal samples we collected from captive rhesus monkeys from 4 provinces in China, 3 were tested as positive forShigellaand 13 tested as positive forSalmonellaby LAMP and real-time PCR assay (Table 2, Fig. 1). No nonspecific amplification was observed. The LAMP and real-time PCR assays had the same positive detection rates ofShigellaandSalmonella.
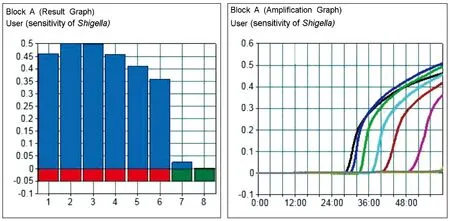
Fig. 6 Sensitivity of the detection of Shigella in LAMP. The minimum detectable limit for Shigella was 10-5 (DNA concentration was approximately 10 pg/μL). CH1: 1.0; CH2-CH8: 1.0×10-1-1.0×10-7.
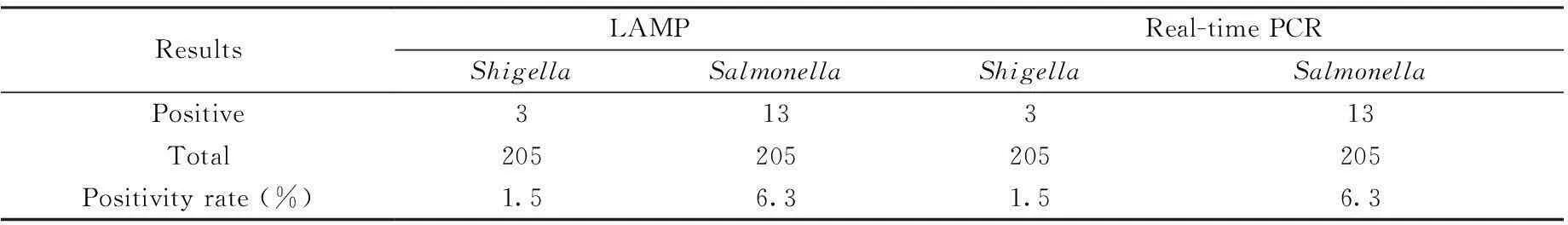
ResultsLAMPReal-timePCRShigellaSalmonellaShigellaSalmonellaPositive313313Total205205205205Positivityrate(%)1.56.31.56.3
Note. LAMP: loop-mediated isothermal amplification. PCR: polymerase chain reaction
3Discussion
ShigellaandSalmonellaare the major pathogens leading to diarrhea in rhesus monkeys. These two bacteria are commonly detected using culturing, PCR,7and enzyme-linked immunosorbent assay8techniques, among others. However, culture requires 2-3 days, and PCR requires specific equipment and >2 h for detection. As such, these methods cannot rapidly detect bacterial diseases in laboratory animals. In contrast, LAMP, a new nucleic acid amplification reaction technique, is advantageous due to its fast and simple reaction conditions used for pathogen detection. In our study, results could be determined after the bacterial nucleic acids were allowed to react in this system at 63℃ for 1 h. The LAMP approach is much faster than the traditional methods.
Most of the currently established LAMP reaction methods determine results using images from the UV analysis of agarose gel electrophoresis.9Furthermore, only the final LAMP reaction result is analyzed, and the methods carry a risk of aerosol contamination in the laboratory. Given the lack of real-time reaction monitoring, eliminating these interfering factors is quite difficult. In this study, we established a real-time LAMP method for detection ofShigellaandSalmonella. The reaction progress was analyzed using a loopamp® real-time turbidimeter LA-320C (Eiken Chemical Co. Ltd., Tokyo, Japan). The instrument be used to automatically observe the reaction in real-time and exclude false-positive and non-specific reaction interference factors according to set analytical standards. In a sample analysis, when the reaction progressed to 24 min, generation of a precipitate was detected (Fig. 5). Analysis of the curves showed that the reaction peaked at 0.5, more than the default of 0.1 positive determination standards. During the efficient LAMP reaction, the amplified target sequence amount was enriched to a critical value and entered a higher reaction rate to achieve the reaction peak. Dilution of the template affected only the reaction starting time and not the reaction efficiency. Therefore, the sample reaction time was associated with the initial amount of DNA template used.
The lowest detection limit of PCR is 200 pg/μL commonly. However, in our study, the detection limits ofShigellaandSalmonellawere both 10 pg/μL, which indicated that the LAMP method had a higher sensitivity than PCR. In examination of the 205 fecal samples, the positive LAMP and PCR rates forShigellaandSalmonellawere the same. These findings suggest that the LAMP method has the same sensitivity as real-time PCR for detectingShigellaandSalmonella, especially the former could be judged with eyes.
The two bacteria real-time LAMP detection methods established in this study are fast (completed within 1 h), sensitive (detection limit of approximately 10 pg/μL), with simple equipment requirements, and have simple experimental operation. In particular, this method has no side effect on animals and required only fecal sample collection. Our findings suggest that the LAMP method is properly applied for the diagnosis of the diarrhea pathogens in the rhesus monkeys. The LAMP primers designed in this assay are applying patents, and the development of a quick kit for macaque diarrhea detection is underway. This investment is scientific and economic for the macaque health maintaining and for the monkey farms.
参考文献
[1]Lu CP. Veterinary Microbiology. (4th ed.): Chinese Agriculture Publisher, 2007.(in Chinese)
[2]Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA [J]. Nucleic Acids Res, 2000, 28: E63.
[3]Zhu H, Lv JZ and Fan F. Development of loop-mediated isothermal amplification (LAMP) method for detection of E. coli O157:H7 [J]. J Mol Diagn Ther 2010, 2: 98-101.
[4]Wang ZR, Chen CF, Yang MF, et al. Development of multi-site loop-mediated isothermal amplification (LAMP) for rapid detection of Mycobacterium tuberculosis [J]. Chin J Prevent Vet Med, 2010, 4: 009. (in Chinese)
[5]Chen S, Ge B. Development of a toxR-based loop-mediated isothermal amplification assay for detecting Vibrio parahaemolyticus [J]. BMC Microbiol 2010; 10: 41.
[6]Mori Y, Nagamine K, Tomita N, et al. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation [J]. Biochem Biophys Res Commun 2001; 289: 150-154.
[7]Soria MC, Soria MA, Bueno DJ, et al. Comparison of 3 culture methods and PCR assays for Salmonella gallinarum and Salmonella pullorum detection in poultry feed [J]. Poult Sci, 2013, 92: 1505-1515.
[8]Cho IH, Irudayaraj J. In-situ immuno-gold nanoparticle network ELISA biosensors for pathogen detection [J]. Int J Food Microbiol, 2013, 164: 70-75.
[9]Okamura M, Ohba Y, Kikuchi S, et al. Loop-mediated isothermal amplification for the rapid, sensitive, and specific detection of the O9 group of Salmonella in chickens [J]. Vet Microbiol, 2008, 132: 197-204.
[收稿日期]2015-09-09
Doi:10.3969/j.issn.1005-4847.2016.01.002
【中图分类号】Q95-33
【文献标识码】A
【文章编号】1005-4847(2016) 01-0007-07
Corresponding author:BAI Jie-ying, E-mail: baijieying@126.com
[作者简介]叶莉(1988-),女,研究实习员,研究方向:人兽共患病; 吴芳(1992-),女,本科生。△本文共同第一作者。[通讯作者]白杰英(1977-),男,副研究员,硕士生导师,研究方向:人兽共患感染性疾病致病机理, E-MAIL: baijieying@126.com; 张小飞(1978-),女,副研究员,研究方向:兽药安全与检测, E-MAIL: wangyizxf@163.com。
[基金项目]十二五后勤科研项目 (CWS11J092);国家自然科学基金 (31272385);十二五重大专项(2011ZXJ09201-031);北京市自然科学基金(5152023)。
