丹参果糖磷酸酶基因的克隆和功能研究
2014-08-06谭何新第二军医大学药学院药用植物学教研室上海200433
谭何新, 刘 颖, 张 磊 (第二军医大学药学院药用植物学教研室, 上海 200433)
1 Introduction
Fructose bisphosphate aldolase (FBA, EC 4. 1. 2. 13) is a key enzyme of the Embden-Meyerhof-Parnas (EMP) pathway of glucose dissimilation[1]. It catalyzes the reversible cleavage of fructose bisphosphate into glyceraldehyde-3-phosphate (G-3-P) and dihydroxyacetone phosphate (DHAP), and is thus important in glycolysis and gluconeogenesis. It can cleave fructose-1-phosphate, which is of significance in fructose metabolism. It also participates into Calvin cycle and continuously catalyzed the rapid and reversible ketose-aldose isomerization of dihydroxyacetone phosphate (DHAP) andD-glyceraldehyde-3-phosphate (G-3-P), followed by the reversible aldol condensation of DHAP and G-3-P to fructose-1,6-bisphosphate (FBP) (Figure 1). Subsequently, the phosphoester bond on C1 of FBP is irreversibly cleaved by fructose-1,6-bisphosphatase to yield fructose-6-bisphosphate (F-6-P)[2]. Aldolases are the most important enzymes for the synthesis of monosaccharides and related compounds[3].
There are two types of aldolases, designated as class I and class II (Figure 1). They present in both prokaryotic and eukaryotic organisms[4], and exhibit no sequence homology[5,6]. The class I aldolases are tetramers and generally found in plant and animal tissues. The aldolase of higher plants belong to the class I type enzymes. This class of aldolase catalyzes the reaction by the Schiff base mechanism[7]. Additionally, some class I aldolases identified in hyperthermophilic archaeaThermoproteustenaxandPyrococcusfuriosusshow no significant DNA sequence similarity when compared with the classical class I enzymes, a new sub-class name, fructose-1,6-bisphosphate aldolase class I A, are proposed for these enzymes[8]. The most thoroughly studies class I aldolase is fructose-1,6-bisphosphate aldolase from rabbit muscle. This tetrameric enzyme with a molecular mass of about 170 ku accepts several aldehydes as substrate[9].
Plant and animal aldolase isoenzymes differ in their expression and compartmentation patterns. Higher plants contain two isoenzymes, one in the cytosol and the other in the chloroplast[10]. In photosynthetic organisms, light activities various signal transduction pathways regulating the growth rate, the expression of genes involved in various metabolic processes are regulated. In higher plants, various phenomena concerning photogermination, phototropism and photoperiodism, including flower initiation, are regulated by light as a signal via phytochromes or other photoreceptors[11]. Animals have three class I isoenzymes, classified as A, B and C, which are expressed in the cytosol of muscle, liver and brain tissue, respectively[12,13]. Fructose bisphosphate aldolase was further characterized as a typical class I enzyme.
SalviamiltiorrhizaBunge is a well-known Chinese herb; its roots have been widely used for the treatment of menstrual disorders and cardiovascular diseases. In this study, we hope to useS.miltiorrhizato extend our knowledge on Embden-Meyerhof-Parnas (EMP) pathway. And we first report the cloning and characterization of a novel fructose bisphosphate aldolase (SmFBA) fromS.miltiorrhiza. Comparison of these reported FBA gene sequences from different species revealed thatFBAgenes are structurally conserved, and might possess similar functions. The work also aims to examine its transcription profile in different tissues and under induction treatments. The successful isolation of theSmFBAgene will be helpful for studying EMP pathway in the near future.
2 Materials and methods
Plantmaterials
Seeds ofS.miltiorrhizawere purchased from local market.The plant was grown in the gardens of Second Military Medical University, Shanghai, China, and identified by Professor Hanming Zhang. Seeds ofS.miltiorrhizawere pretreated with 75% alcohol for 1 min, washed three times with distilled water, followed by the treatment of 0.1% HgCl2for 5 min and by four rinses with sterile distilled water. The sterilized seeds were then incubated between several layers of sterilized wet filter paper and cultured on MS basal medium for germination. The seedlings were grown at 25 ℃ under 12 h light/ 12 h dark photoperiod cycles for 2 months until inducing treatment and RNA isolation.
RNAandDNAisolation
Total RNAs of various treatments from 2-month-oldS.miltiorrhizaplant were extracted using TRIzol reagent (GIBCO BRL) according to the manufacturer′s instruction[16]. The genomic DNA ofS.miltiorrhizawas isolated using a Cetyl trimethyl ammonium bromide (CTAB)-based method. The quality and concentration of RNA and DNA samples were examined by EB-stained agarose gel electrophoresis and spectrophotometer analysis before use.
Cloningofthefull-lengthcDNAencodingSmFBA
For 3′ RACE, cDNA synthesis was implemented by using the 3′ RACE system. An aliquot of isolated 100 ng total RNA extracted from leaves was reversely transcribed with a cDNA synthesis strategy named “First-stand cDNA synthesis” by using Clontech SmartTM RACE cDNA amplification kit (BD Biosciences, USA).SmFBAspecific 3′ primers Sm3 (5′-GGTGCCAATGAGCCATCACAG-3′), Sm 3-Nested (5′-GGTGACAGAGCGTGTTCTTGC-3′) were designed and synthesized according to the conserved sequence among the knownFBAs(BAE48790, CAB77243, NP_568127). Sm3 and UPM (provided with in the kit) were used for the 3′ RACE-PCR and cDNA was used as template. PCR amplification was performed under the following conditions: cDNA was denatured at 94 ℃ for 3 min followed by 35 cycles of amplification (94 ℃ for 1 min, 58 ℃ for 1 min, 72 ℃ for 1 min and 30sec) and by 72 ℃ for 10 min. An aliquot of 0.5 μl of the first round PCR product was subsequently used as template for nested PCR amplification using primers Sm3-nested and UPM under the following PCR condition: 94 ℃ for 3 min followed by 35 cycles of amplification (94 ℃ for 1 min, 58 ℃ for 1 min, 72 ℃ for 1 min and 30 sec) and by 72 ℃ for 10 min. The amplified PCR product was purified and cloned into pMD 18-T vector (Takara, Japan) followed by sequencing.
For 5′ RACE,SmFBAspecific 5′ primers Sm5 (5′-GAAGAAGATGGCGTTGCCGAA-3′) was designed according to the conserved sequence among the knownFBA, and used to amplify the 5′ end ofSmFBAcDNA. For 5′ RACE, 100 ng total RNA was reversely transcribed by “First-stand cDNA synthesis” by using Clontech SmartTM RACE cDNA Amplification Kit (BD Biosciences, USA). The first round of PCR was performed with primers Sm5 and UPM (provided by the kit) using 2.5 μl First-strand cDNA as template in a total volume of 50 μl reaction mixture under the following PCR condition: cDNA was denatured at 94 ℃ for 3 min followed by 35 cycles of amplification (94 ℃ for 1 min, 58 ℃ for 1 min, 72 ℃ for 1 min and 30 sec) and by 72 ℃ for 10 min. The amplified 5′ PCR product was purified and cloned into pMD 18-T vector (Takara, Japan) followed by sequencing.
For full-length cDNA sequence ofSmFBA, by comparing and aligning the sequences of 3′ and 5′ PCR products, specific primers FBA-F (5′- ATGACTGCCTACCGCGGAAAG-3′) and FBA-R (5′- TTATGCTCTCTCCTCCAACCG- 3′) were designed. The ORF sequence was obtained through PCR reaction by using first-strand cDNA as the template under the following PCR condition: 94 ℃ for 3 min followed by 35 cycles of amplification (94 ℃ for 1 min, 58 ℃ for 1 min, 72 ℃ for 1 min) and by 72 ℃ for 10 min. The PCR product was purified and cloned into pMD 18-T vector followed sequencing.
Bioinformaticsanalysisandphylogeneticconstruction
ORF translation and molecular mass calculation of the predicted protein were carried out on Vector NTI Suite 11. GenBbank BLASTs were carried out on NCBI (http://www.ncbi.nlm.nih.gov/). Phylogenetic analysis of SmFBA and other known FBAs from other plant species retrieved from GenBank were aligned ClustalX software (version 1.80) and subsequently a phylogenetic tree was constructed by the neighborjoining (NJ) method using the software of MEGA 2.0[17-19].
Southernblotanalysis
Aliquots of genomic DNA (15 μg/sample) were digested overnight at 37 ℃ - withBglII,EcoRI,HindIII,SacIrespectively, which did not cut within the full-length cDNA ofSmFBA. The digested DNA was fractionated by 1.0% agarose gel electrophoresis, transferred onto a positively charged hybond-N+nylon membrane (Amersham Pharmacia, UK) and hybridized with the biotin-labeled 3′ region coding sequence ofSmFBA(500 bp) as the probe, which was derived from the cDNA sequence. Probe labeling (biotin), hybridization and signal detection were performed using Gene Images Random Prime Labeling Module and CDPStar Detection Module following the manufacturer′s instructions (Amersham Pharmacia, UK). The hybridized signals were visualized by exposure to Fuji X-ray film at room temperature for 30 min.
ExpressionprofileofSmFBAindifferenttissues
Total RNA was reversely transcribed by using AMV reserve transcriptase (Takara, Japan) to generate cDNA. Gene specific primers (5′-ACGTTATGCCATCATCTGCCA-3′ and 5′-GCCTCTTCCTCACTCTGTCCA-3′) were designed according to the corresponding sequences ofS.miltiorrhiza. Partial of polyubiquitin gene was amplified with primers (5′-ACCCTCACGGGGAAGACCATC-3′ and 5′-ACCACGGAGACGGAGGACAAG-3′) as a control. Real-time PCR was performed according to manufacturer′s instruction (Takara, Japan) under the following condition: 1 min pre-denaturation at 95 ℃, 1 cycle; 10 s denaturation at 95 ℃, 20 s annealing at 64 ℃, 15 s collection fluorescence at 72 ℃, 40 cycles. The products of real-time quantitative PCR were run on 1.5% agarose gel electrophoresis and showed an equal-sized band as predicted. Quantification of the gene expression was done with comparative CT method. Experiments were performed in triplicate.
Inductiontreatments
S.miltiorrhizaseedlings were grown on SM medium for 30 days (25 ℃, 10 h light/14 h dark). Leaves were sprayed with solution of 100 μmol/L MeJA (Dissolved in dimethyl sulfoxide)、and 100 μmol/L GA3(GA3was dissolved in small amount of ethanol, than metered volume by water) and 0.5 mol/L NaCl (dissolved in water), respectively.
TheexpressionofrecombinantSmFBAinE.coli
The full length SmFBA cDNA was cloned into pET28a vector (Novagen) byEcoRI andXhoI restriction sites, and the constructed pET-SmFBA was introduced intoE.colistrain BL21 by electroporation. 1 ml of overnight-cultured transformant was inoculated into 100 ml of fresh LB medium with 50 mg/L kanamycin at 37 ℃. When the OD600reached 0.6, 0.5 mmol/L iso-propyl thiob-D-galactoside (IPTG) was added and incubation at 37 ℃ 6 h.
EnzymeassayofrecombinantSmFBA
D-fructose-1,6-phosphate (FDP) have high specialization for aldolase, 2,4-dinitrophenylhydrazine (DNPH) may react with dihydroxyacetone phosphate (DAP) derived from decomposition product of FDP to form phenylhydrazone. Ultraviolet wavelength at 540 nm can be used to measure the product absorption value[20]. Enzyme assay at various reaction times over the range 0-5 min were performed in 2 ml mixture [50 mmol/L Tris-HCl buffer pH 7.4, 4 mmol/L FDP trisodium salt (FDPNa3), 5 mmol/L DNPH]. The reaction was allowed to proceed at 37 ℃ after a suitable aliquot of the enzyme was added. Then, the optical density at 540 nm was monitored once every 1 min by spectrophotometric measurement.
3 Results
Isolationandcharacterizationofthefull-lengthcDNAofSmFBAgene
Using the RACE method and primers mentioned in the methods part, cDNA fragments of 559 bp and 708 bp were amplified by 3′ RACE and 5′ RACE respectively, a core region of 883 bp was also amplified by RT-PCR. The three sequence above were assembled with Vector NTI Suite 11 and the full-length SmFBA cDNA (GenBank accession no. FJ540907) was subsequently amplified by proof-reading PCR amplification with primers mentioned above. The full-length cDNA of SmFBA was 1 390 bp and contained a 1 065 bp ORF which encoding a 355-amino-acid protein, flanked by stretches of 112 bp and 213 bp at the 5′-untranslated and 3′-untranslated regions with 24 bp poly A tail. BLAST search revealed that the nucleotide sequence ofSmFBAhad sequence similarities toFBAsfrom other species.
Generationandcharacterizationofthefull-lengthDNAofSmFBA
Gene-specific primers derived from the start and stop codon regions of theSmFBAcDNA were designed and synthesized to isolate the genomicSmFBAgene by PCR. The PCR for genomic sequence resulted in a clear band of 2 101 bp, which was 1 036 bp longer than that of the coding sequence. The comparison with the cDNA showed that the genomic DNA and cDNA matched base to base except that the genomic DNA contained two introns. The lengths of the three exons were 27 bp, 230 bp and 980 bp respectively. The lengths of the two introns were 852 bp and 105 bp respectively (Figure 2A). It was also found that the putative splicing site obeyed the GT/AG rule.
CharacterizationofthededucedSmFBA
By using the software of pI/MW tool (www.expasy.org), the calculated isoelectric point (pI) and molecular weight of the deducedSmFBAwere predicted to be 5.60 and 37.78 ku. Analysis of the secondary structure of the putative protein sequence ofSmFBAindicated that the secondary structure of theSmFBAis mainly consisted of α-helix (52.30%) and random coils (27.20%), while extended strand (12.97%) and β-turn (7.53%) contribute a little. Protein-protein BLAST showed that on the amino acid levelSmFBAhad high homology to FBAs from other plant species (Figure 2B). Through Vector NTI Suite 11.0 full-length alignment results showed thatSmFBAshared 74.1% identity to LeFBA from Lycopersicon esculentum, 75.2% identity to AtFBA fromArabidopsisthaliana, and 53.7% identity to OsFBA fromOryzasativarespectively. All of these FBAs have four predicted N-myristoylation sites and a class I FBA active site (http://mendel.imp.ac.at/myristate/SUPLpredictor.htm). These results revealed thatSmFBAhas a high degree of similarity to other FBAs, and we presumed they executed the same function in plant metabolism.
Molecularevolutionanalysis
FBA is a part of the EMP biosynthetic pathway. It exists in many species and contributes in synthesis of varied natural products. In order to study the evolutionary relationships among different FBA proteins from various plant species, a phylogenetic tree was constructed using the MEGA program based on the deduced amino acid sequences of predictedSmFBAand other FBA proteins from other species, from prokaryote to eukaryote, from monocot to dicot plants (Figure 2C). FBA sequence formed several distinct species-specific clusters. For example, all monocot plants formed a cluster, FBA from the prokaryote species was separated from other eukaryote species. According to the phylogenetic tree,SmFBAgot the closest evolutionary relationship with another well-known plant speciesP.americanaandA.thaliananext.

Figure 2 Bioinformatics analysis of SmFBA sequence
A.Exon and intron analysis of FBA inS.miltiorrhizagenome. Black background indicate intron, white background indicate exon respectively.
B.Multiple alignments,andC.Phylogenetictree analysis of fructose-bisphosphate aldolases isolated from various plants. FourN-myristoylation sites and one class-I FBA active site were signed. The GenBank accession numbers are as follows:Salviamiltiorrhiza(SmFBA, FJ540907);Codonopsislanceolata(ClFBA, BAE48790);Perseaamericana(PaFBA, CAB77243);Oryzasativa(OsFBA, CAA37290);Glycinemax(GmFBA, AAR86689);Solanumtuberosum(ABB29926);Populustrichocarpa(BK96406);Arabidopsisthaliana(NP_568127);Zeamays(CAA31366);Sesuviumportulacastrum(ACG68894);Physcomitrellapatenssubsp.Patens(XP_001768658);Schistosomabovis(ACC78612);Homosapiens(CAA30270);Musmusculus(NP_031464);Oryctolaguscuniculus(NP_001075707);Macacamulatta(XP_001108059);Agrobacteriumtumefaciensstr. C58(NP_356881);Legionellapneumophilastr.Corby(YP_001252121);MethanocorpusculumlabreanumZ(YP_001030041);BacillushaloduransC-125 (BAB06033);Campylobacterjejunisubsp.JejuniCG8421(ZP_03222218).
SouthernblotanalysisofSmFBAinS.miltiorrhizagenome
To examine the copy number of theSmFBAgene, 15 μg genomic DNA ofS.miltiorrhizawas digested withBglII,EcoR V,HindIII andSacI, respectively. The result showed that one or two bands were found in each lane (Figure 3), suggesting thatSmFBAgene was a low-copy number gene in S. miltiorrhiza genomic.
Tissue-specificandinducedexpressionprofileofSmFBA
The expression pattern of genes in different tissues reflects the distribution of metabolites[14]. To investi-gate theSmFBAexpression pattern in different tissues ofS.miltiorrhiza, total RNA was extracted from roots, stems, and leaves, respectively, and subjected to real-time PCR analysis. The result showedSmFBAexpression could be detected in all tissues, suggesting thatSmFBAa constitutively expressed but at different expression levels in various tissues.SmFBAexpressed strongly in roots and stems, weakly in leaves (Figure 4A). To characterize the transcription pattern ofSmFBAunder different elicitors′ induction during the culture period,S.miltiorrhizaseedlings were treated with MeJA, GA3and NaCl, which are well known significant regulator in the biosynthesis of secondary metabolites in plants, and were harvested for RNA isolation after 24 hours. As shown in Figare. 4B, the expression ofSmFBAwas significantly up-regulated upon treatments. After 24 h treatment, the transcription value ofSmFBAincreased 1.8-, 5.7-, 4.5-fold higher than the wild-type control, under the induction of MeJA, GA3and NaCl respectively. It obviously showed that elicitors promote the expression ofSmFBA.
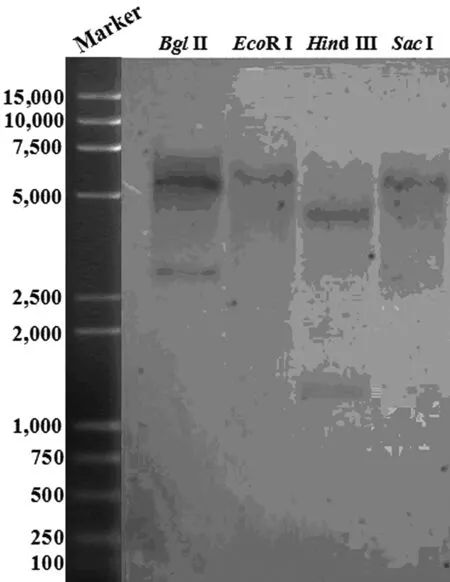
Figure 3 Southern blot analysis of SmFBA gene
Genomic DNA (15 μg /lane) is digested with Bgl II,EcoRI,HindIII andSacI respectively, followed by hybridization with 3′ UTR ofSmFBAsequence as the probe.

Figure 4 Expression profiles of SmFBA determined by qRT-PCR
A.Expression patterns ofSmFBAin different organs.
B.Induction ofSmFBAexpression upon treatments with methyl 0.1 mmol/L jasmonate, 0.1 mmol/L GA3and 0.5 mmol/L NaCl for 6 hours.Data are presented as relative to untreated control plants (set as 100%) (Means±SE;n=3 biological replications)
RecombinantSmFBAhaveenzymeactivityandcouldimprovethesalt-toleranceofE.coli
RecombinantSmFBAwas rapidly and largely induced inE.coliby IPTG. SDS-PAGE analysis showed the recombinantSmFBAwas about 41ku (Figure 5A), which was consistent with the predicted value. The enzyme activity of recombinantSmFBAinE.coliwas detected as shown in Figure 5B. With the increase of reaction time, the yielded products of recombinantSmFBAshowed an upward trend. While, the yielded products of empty pET28a vector and boiled recombinantSmFBAwere not obvious. The result demonstrated that recombinant proteinSmFBAhas activity inE.coli. In addition, we also test the NaCl stress tolerance of theE.coliexpressingSmFBA, and found its NaCl tolerance improved greatly compared with the wild-typeE.colior theE.coliexpressing empty vector (Table 1).
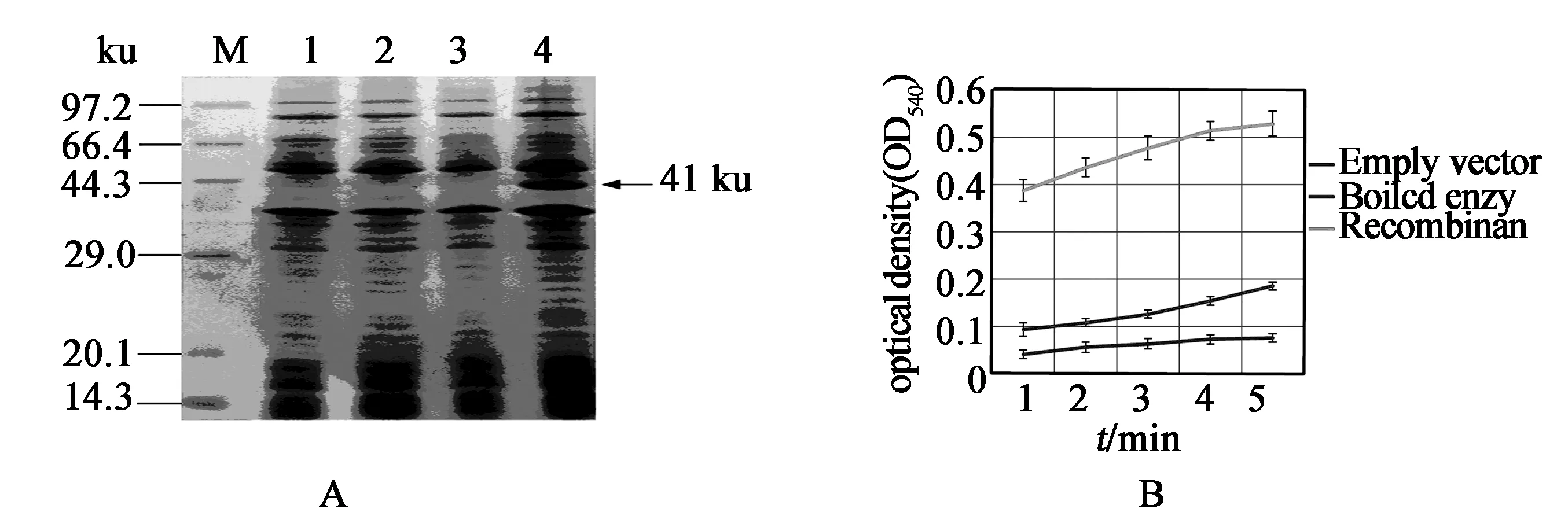
Figure 5 SDS-PAGE analysis of pET directional TOPO-SmFBA fusion protein in E. coli
Table1GrowthofE.coliunderNaClstress*

VariableWild typeEmpty vectorSmFBASlope0.0520.0480.083
*Slope is measured as change in OD600per hour. All strains were grown in LB liquid medium containing 0.5 M NaCl at 37 ℃ for 5 hours (transformants plus kanamycine).
4 Discussion
In this paper, the full-length cDNA and genomic DNA sequences ofSmFBAgene were isolated fromS.miltiorrhiza, revealing that theSmFBAgene contained two introns. The deducedSmFBAprotein showed high identity to FBA proteins from other plant species via multiple alignments, implied thatSmFBAmight have the same catalytic function as other FBAs. Previous studies have shown that FBA belongs to a multigene family in some plant genome, for example, there were at least seven FBA-like genes inZeagenome(Genbank accession no. AF338139, AF466202, AF466203, AF464738, AF466646, AF528565, EF445629). Southern-blot analysis revealed thatSmFBAgene was a low-copy number gene in theS.miltiorrhizagenomic. The topology of the phylogenetic tree is generally in good agreement with the traditional taxonomy classification with three main braches. The plant FBAs, animal FBAs and bacteria FBAs were grouped into three clusters, respectively. According to the phylogenetic tree,SmFBAgot the closest evolutionary relationship with another well-known plant speciesP.americanaandA.thaliananext.
S.miltiorrhizais a commonly used traditional Chinese medicine for improving body function(e.g. promoting circulation and improving blood flow), as well as for the treatment of angina pectoris, myocardial infarction and other cardiac symptoms[15]. The active components ofS.miltiorrhizacould be classified as lipid-soluble and water-soluble ones. In recent years, more and more persons do research in secondary metabolic pathway of lipid-soluble and water-soluble active components because of the importance of active components. Few people pay close attention to primary metabolic inS.miltiorrhiza. We studied the role of FBA inS.miltiorrhizaand do analysis of bioinformatics for the first time.
In summary, we successfully isolated and characterized the full-lengthSmFBAcDNA from the medicinal plant S. miltiorrhiza. The cloning and characterization ofSmFBAwill be helpful to understand more about the role of FBA and enable us to penetrate deeply its role in photosynthesis at the molecular level.
【参考文献】
[1] Siddiqui KA, Banerjee AK. Fructose 1,6-bisphosphate aldolase activity ofRhizobiumspecies[J]. Folia Microbiol,1975, 20: 412.
[2] Fridlyand LE, Scheibe R. Regulation of the Calvin cycle for CO2fixation as an example for general control mechanisms in metabolic cycles[J]. BioSystems,1999, 51: 79.
[3] Jones JB. Enzymes in organic synthesis[J]. Tetrahedron Lett,1986, 42: 3351.
[4] Rutter WJ. Evolution of aldolase[J]. Fed Proc,1964, 23: 1248.
[5] Marsh JJ, Lebherz HG. Fructose-bisphosphate aldolases: an evolutionary history[J]. Trends Biochem Sci,1992, 17: 110.
[6] Doolittle RF. Convergent evolution: the need to be explicit[J]. Trends Biochem Sci,1994, 19: 15.
[7] Perham RN. The fructose 1,6-bisphosphate aldolases: same reaction, different enzymes[J]. Biochem Soc Transcript,1990, 18: 185.
[8] Siebers B, Brinkmann H, Dorr C,etal. Archaeal fructose-1,6-bisphosphate aldolases constitute a new family of archaeal type class I aldolase[J]. J Biol Chem,2001, 276: 28710.
[9] Effenberger F, Straub A. A novel convenient preparation of dihydroxyacetonphosphate and its use in enzymatic aldol reactions[J]. Tetrahedron Lett, 1987, 28: 1641.
[10] Anderson LE, Advani VR. Chloroplast and cytoplasmic enzymes. Three distinct isoenzymes associated with the reductive pentose phosphate cycle[J]. Plant Physiol,1970, 45: 583.
[11] Briggs WR, Christie JM. Phototropins 1 and 2: versatile plant blue-light receptors[J]. Trends Plant Sci,2002, 7: 204.
[12] Joh K, Arai Y, Mukai T,etal. Expression of three mRNA species from a single rat aldolase A gene, differing in their 5′ non-coding regions[J]. J Mol Biol,1986, 190: 401.
[13] Maire P, Gauttrons S, Hakim V,etal. Characterization of three optional promoters in the 5′-region of the human aldolase A gene[J]. J Mol Biol,1987, 197: 425.
[14] Kai GY, Jiang JH, Zhao DL,etal. Isolation and expression profile analysis of a new cDNA encoding 5-alpha-taxadienol-10-beta-hydroxylase from Taxus media[J]. J Plant Biochem Biotechnol,2006, 15: 1.
[15] Zhou LM, Zuo Z, Moses S. Danshen: an overview of its chemistry, pharmacology, pharmacokinetics, and clinical use[J]. J Clin Pharmacol,2005, 45: 1345.
[16] Jaakola L, Pirttila AM, Halonen M,etal. Isolation of high quality RNA from bilberry (VacciniummyrtillusL.) fruit[J]. Mol Biotechnol,2001, 19 : 201.
[17] Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees[J]. Mol Biol Evol,1987, 4: 406.
[18] Thompson JD, Higgins DG, Gibson TJ. Clustal-W-improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice[J]. Nucl Acids Res,1994 22: 4673.
[19] Kumar S, Tamura K, Jakobsen IB,etal. MEGA2: molecular evolutionary genetics analysis software[J]. Bioinformatics,2001, 17, 1244.
[20] Fan W, Zhang Z, Zhang Y. Cloning and molecular characterization of fructose-1,6-bisphosphate aldolase gene regulated by high-salinity and drought inSesuviumportulacastrum[J]. Plant Cell Rep,2009, 28(6):975.
