Analytical characteristics of a qPCR-based molecular diagnostic assay-conceptual considerations for laboratory personnel
2014-04-18TanujShukla,PrabhakarRaoKaipa,ShesheerKMunpally等
Analytical characteristics of a qPCR-based molecular diagnostic assay-conceptual considerations for laboratory personnel
Dear Editor:
Quantitative real-time PCR has revolutionized molecular diagnostics with its ease of use,increased sensitivity and specificity and low turnaround time[1]. PCR/quantitative PCR(qPCR)-based assays offer a distinct advantage over other serological/conventional diagnostic approaches[7-10].The ability to diagnose infectious diseases has benefited from the availability of US FDA approved and Conformite Europeenne (CE)-marked qPCR-based in-vitro diagnostic kits from international companies.The high-quality kits are calibrated with the World Health Organization(WHO) reference standards and the National Institute for Biological Standards and Control(NIBSC)standards. They are tested for proficiency using the College of American Pathologists(CAP)guidelines and Quality Control for Molecular Diagnostics(QCMD).In addition to essential components such as qPCR mix,primer-probe mix and quantitation standards,these kits are also provided with nucleic acid extraction systems, internal controls for the quality check on nucleic acid extraction procedures,and calibrators with defined values for the evaluation of results.The reporting scale is also slowly shifting from copies/mL to international unit(IU)/mL,which provides more precise and accurate results.A process overview with the steps involved in a qPCR-based molecular diagnostic assay is presented inFig.1.Possible technical deficiencies that may affect assay performance and lead to false results include: inadequate sample storage,the preparation and quality of nucleic acid,poor choice of reverse-transcription primers and probes for the PCR,and inappropriate data and statistical analyses[1].Another major concern for a molecular biologist/laboratory technician while using these qPCR-based kits in a laboratory is the limit of detection(LOD)/analytical sensitivity;two other frequently asked questions regard assay specificity and broad linear dynamic range.Finally,an important aspect is the interpretation of results,which is critical for accurate reporting.
The primary requirements for an ideal molecular diagnostic laboratory are optimal conditions and proper segregation.To accomplish this,the sample extraction and pre-and post-PCR analysis areas should be segregated. The workflow should be unidirectional,with efficient cleaning and fumigation procedures that help prevent cross contamination and the tedious process of repeating tests.In addition,biosafety laminar air flow units,calibrated micropipettes,the use of aerosol barrier tips, appropriate room temperatures,and sample storage facilities are essential for maintaining proper quality control[5]. The analysts responsible for performing the experiments should be trained in GLP practices.Before performing the experiment,the user should thoroughly read and understand the instructions provided in the kit manual. Some manufacturers sell machine specific kits that are not validated for all thermal cyclers;it is advised to check the manufacturer specifications before procuring the kit.
The second consideration is the quality of the sample used for extraction of nucleic acids.The nucleic acid content varies depending on the sample source (blood,body fluids,respiratory samples,and body excretions)and improper storage conditions increase the chances of inhibitors,microbial contamination, and nucleic acid degradation.Therefore,collection, transport,storage and processing time become very critical to achieving optimal results.The Clinical and Laboratory Standards Institute(CLSI)provides guidelines for a diagnostic laboratory and specifies,in detail, every aspect of clinical sample handling[5].For an accurate diagnosis,the purity of nucleic acid is very important.Extreme care should be taken while extracting DNA/RNA.The yield of nucleic acid varies depending upon the extraction system as well as the source of the sample used in the individual laboratory. Recently,technologies in the extraction systems ofnucleic acids have drastically improved,which are based on magnetically charged silica beads and affinity membrane based columns[5].The qPCR diagnostic kits are validated on one of these or combinations of different extraction systems and the same extraction systems are recommended by the kit manufacturer.It is advised that the nucleic acid extraction procedures be performed in a BSL3 cabinet with calibrated pipettes and barrier tips which stop sample cross-contamination by preventing aerosols from reaching the barrel of pipette.Many manufacturers provide internal amplification controls that can co-purify and co-amplify with the target nucleic acid.This is useful for identification of template loss and inhibition during sample processing[2].
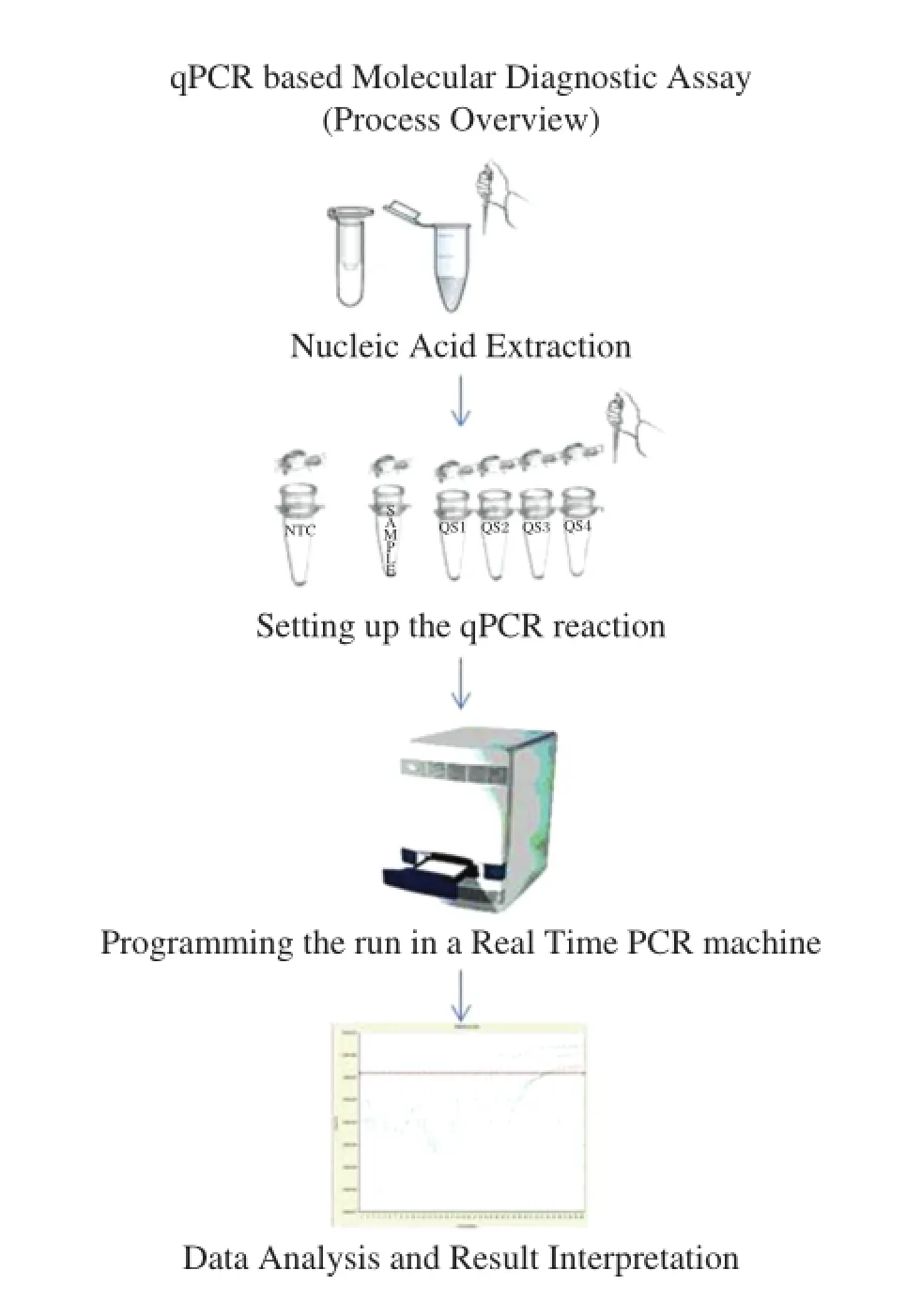
Fig.1qPCR based molecular diagnostic assay(process overview)
The analytical sensitivity of any qPCR kit can be measured by conducting standard runs from known reference standards for the particular assay,which can be procured from NIBSC(WHO).It is important to read the kit manufacturer′s instructions regarding repeatability and reproducibility of the assay.The analytical sensitivity of the assay is largely dependent on the extraction system,more precisely the sample volume taken for extraction and the volume used for the elution of nucleic acid target.Therefore,it is important for the end user to exactly follow the instructions provided by the manufacturer;otherwise,deviation from the recommended protocol may introduce inconsistencies in reporting.However,a proper validation study while manipulating with the sample extraction volumes can improve the limit of detection to a certain extent.In some cases,concentrating the sample before nucleic acid extraction helps improve the analytical sensitivity by approximately tenfold[3].
An analyst should run all the quantitation standards/ calibrators provided in the kit along with serial dilutions of samples/standards with known values during the first run to obtain linear quantitation range for the assay.An accurate and better linear quantitation range can only be estimated/cross verified when the assay slope is between-3.1 to-3.6,the assay efficiency is from 90%to 110%and y intercept close to 40 Ct. The use of efficient hot start polymerases,thermostable reverse transcriptases,and optimized primer-probe concentrations/compositions along with rigorous precision analysis by the kit manufacturers helps obtain a broad linear quantitation range.
Because most of these kits use highly specific hydrolysis probes[5],extensive clinical evaluation,along with validation with international reference panels,has to be included to rule out the possibility of low specificity. Alternatively,the specificity can be cross checked using international reference panels and by using a BLAST search based approach[4].The analyst is also advised to prepare a master mix of all reagents and then dispensing them into respective tubes,including NTC,prior to opening clinical samples and standards.This ensures uniform distribution and reduces chances of cross contamination from positive controls/samples.
An analyst should always consider the variables introduced during the run by comparing the Ct values from the original run with all standards/calibrators performed and the consecutive runs with at least 3 standards.This ensures consistency in assay performance over the period of time/runs.Sometimes,due to manual error or degradation of quantitation standards,the slope may fall out of range.Omitting the outlier standard and calculating the slope with the remaining standards can correct this.This validates the experiment performed by giving accurate results and prevents repetition.If possible,the calculated values of qStandards must be cross checked with the original run before generating patient report.It is also important to ensure the no-template control does not have any amplification.The notemplate control amplification of Cq>30 suggests the presence of high levels of contamination in the laboratory[1].In this scenario,the laboratory area, including micropipettes and the laminar air flow unit, should be cleaned thoroughly per GLP procedures.Ano-template control run should be performed for the suspected assay after employing cleaning procedure to ensure the area is contamination free.
Most commercially available real time PCR instruments automatically adjust the threshold after completion of run for analysis.However,it is important for the user to adjust the threshold manually above the baseline/negative control while viewing the amplification plot in linear phase and not in log phase,if available. A linear phase view of the quantitation assay gives a precise location or point where the threshold can be adjusted.Importing the standard curve(available in some thermocyclers software)should be avoided and, if required,one of the quantitation standards should be run along with the samples,to assess assay efficiency and quantitation drift.
The necessity for ensuring quality-assurance measures for qPCR-based assays is well recognized.The main advantage of qPCR over conventional PCR assays is accurate quantification of target nucleic acids.The detection mechanism of a real time PCR system also helps to improve the sensitivity of the assay.Thus,the field of molecular diagnostics is moving at a rapid pace, replacing the conventional serological approach. Moreover,it is very important to understand the intricacies of the system and provide accurate diagnosis.The advent of digital PCR promises to provide even higher sensitivity and precision,especially in the diagnosis of complex infectious microorganisms.
Yours Sincerely,
Dr.Tanuj Shukla and Dr.Prabhakar Rao Kaipa
Department of Genetics and Biotechnology,
Osmania University,
Hyderabad 500007,
India.
Dr.Shesheer K Munpally and Dr.Rachana Tripathi
RAS Lifesciences Pvt.Ltd.,
Hyderabad 500007,
India.
Tel:+919704567314;
E-mail:tanuj.aaidu@gmail.com
The authors reported no conflict of interests.
[1] Bustin SA,Benes V,Garson JA,Hellemans J,Huggett J, Kubista M,et al.The MIQE guidelines:minimum information for publication of quantitative real-time PCR experiments.Clin Chem 2009;55:611-22.
[2] Nolan T,Hands RE,Bustin SA.Quantification of mRNA using real-time RT-PCR.Nat Protoc 2006;3:1559-82.
[3] Ferreira-Gonzalez A,Yanovich S,Langley MR, Weymouth LA,Wilkinson DS,Garrett CT.Enhanced analytical sensitivity of a quantitative PCR for CMV using a modified nucleic-acid extraction procedure.J Clin Lab Anal 2000;14:32-7.
[4] Lemmon GH,Gardner SN.Predicting the sensitivity and specificity of published real-time PCR assays.Ann Clin Microbiol Antimicrob 2008;25;7:18.
[5] Espy MJ,Uhl JR,Sloan LM,Buckwalter SP,Jones MF, Vetter EA,et al.Real-time PCR in clinical microbiology: Applications for routine laboratory testing.Clin Microbiol Rev 2006;19:165-256.
[6] Shah M,Chihota V,Coetzee G,Churchyard G,Dorman SE.Comparison of laboratory costs of rapid molecular tests and conventional diagnostics for detection of tuberculosis and drug-resistant tuberculosis in South Africa. BMC Infect Dis 2013;13:352.
[7] Rodrigues C,Deshmukh M,Jacob T,Nukala R,Menon S, Mehta A.Significance of HBV DNA by PCR over serological markers of HBV in acute and chronic patients. Indian J Med Microbiol 2001;19:141-4.
[8] Nilsson AC,Bjo¨rkman P,Persson K.Polymerase chain reaction is superior to serology for the diagnosis of acute Mycoplasma pneumoniae infection and reveals a high rate of persistent infection.BMC Microbiol 2008;8:93.
[9] Cota GF,DeSousa MR,Demarqui FN,Rabello A.The diagnostic accuracy of serologic and molecular methods for detecting visceral leishmaniasis in HIV infected patients:meta-analysis.PLoS Negl Trop Dis 2012;6: e1665.
[10]Gilber SR,Alban SM,Gobor L,Bescrovaine JDO, Myiazaki MI,Thomaz-Soccol V.Comparison of conventional serology and PCR methods for the routine diagnosis of Trypanosoma cruzi infection.Rev Soc Bras Med Trop 2013;46:310-5.
Received 08 March 2014,Revised 14 March 2014,Accepted 25 March 2014,Epub 27 April 2014
ⓒ2014 by the Journal of Biomedical Research.All rights reserved.
10.7555/JBR.28.20140046
杂志排行
THE JOURNAL OF BIOMEDICAL RESEARCH的其它文章
- Standardized training for resident doctors in China
- Metabolic regulation by protein tyrosine phosphatases
- Apolipoprotein B100 quality control and the regulation of hepatic very low density lipoprotein secretion
- A genetic variant in pseudogene E2F3P1contributes to prognosis of hepatocellular carcinoma
- Expression of human hepatic lipase negatively impacts apolipoprotein A-I production in primary hepatocytes from Lipc-null mice
- Class A scavenger receptor activation inhibits endoplasmic reticulum stress-induced autophagy in macrophage
