A Preliminary Phylogenetic Analysis of Luidia (Paxillosida:Luidiidae) from Chinese Waters with Cytochrome Oxidase Subunit I (COI) Sequences
2013-07-29XIAONing1LIURuiyu1YUANShuai3andSHAZhongli1
XIAO Ning1), 2), LIU Ruiyu1), YUAN Shuai3), and SHA Zhongli1), *
A Preliminary Phylogenetic Analysis of(Paxillosida:Luidiidae) from Chinese Waters with Cytochrome Oxidase Subunit I (COI) Sequences
XIAO Ning, LIU Ruiyu, YUAN Shuai, and SHA Zhongli
1),,266071,2),100049,3),041000,
Forbes(Paxillosida: Luidiidae) are common soft bottom sea stars with 49 described species. Because of substantial morphological diversity, the taxonomy of the genus is complex and hasn’t been resolved definitely. In order to resolve general taxonomic issues, and determine species boundaries and phylogenetic relationships within the genus, the sequences of cytochrome oxidase subunit Ι (COΙ) gene from 24 specimens ofbelonging to eight taxa in Chinese waters, were studied. Threesequences of two species in genusfrom GenBank were used to analyze the phylogenetic relationships. The molecular phylogeny exhibited three main clades, each with strong bootstrap support: Clade A includingfrom the Sea of Japan; Clade B including seven nominal species (von Martens,Goto,Liu, Liao and Li,Fisher,Fisher,Sladen andGray) from Chinese waters; and Clade C includingMüller & Troschel from Chinese waters. Our molecular phylogeny results support the morphological Quinaria-Group and Alternata-Group assigned by Döderlein. Seven nominal species we sampled do not exhibit genetic distances that are large enough to recognize them as separate species. Cryptic species may exist in ‘’ from the Yellow Sea and the Sea of Japan. Meaningful morphological characters need further investigation in.
Echinodermata;; China; DNA taxonomy; cryptic species
1 Introduction
The starfish genusForbes, 1839, is the only valid genus of Luidiidae (Asteroidea: Paxillosida) with 49 species described worldwide to date (Mah and Hansson, 2012). It has a wide distribution, mainly in shallow waters of tropical and subtropical seas (Clark and McKnight, 2000), and is partly confined to temperate waters (Djakonov, 1950). These animals mainly feed on molluscs and other echinoderms, live in sandy or muddy substrate (Sloan, 1980). Only a few species are recorded in considerable depths, such asDüben & Koren, 1845 in up to 1300m (Clark and Downey, 1992).is a common inshore species and has been the subject of many biological studies (Schram., 2011; Gui., 2011).
There are many taxonomic studies of. Döderlein (1920) divided more than 40 species ofincluding ten subgenera into four groups. He emphasized the form and appearance of primary ossicle systems and the deve- lopment of spines, spinelets, and pedicellariae as useful morphological features. Blake (1973) studied the internal consistency of Döderlein’s groups with respect to ossicle morphology. The ossicle morphology of most species ofsupports the systematic arrangement by Döderlein (1920). Fell (1963) elevated most of Döderlein’s subgenera to generic rank. Clark & Downey (1992) stated that the limits of the four main groups and some of the subgenera within them were blurred and did not accept Fell’s taxonomic treatment.
In China,von Martens, 1865 andGoto, 1914 were first reported from Qingdao by Chang (1948). From then on six speciesoffrom southern China have been recorded (Liao and Clark, 1995), includingFisher, 1913,Gray, 1840,Sladen, 1889,Müller & Troschel, 1842,Fisher, 1913 andvon Martens. Liu. (2006) described two new species,Liu, Liao & Li, 2006 from the Yellow Sea, andLiu, Liao & Li, 2006 from the South China Sea. To date, nine species ofhave been recorded from Chinese waters (Liu., 2007). However, morphological variation is very high inand taxonomic boundaries are difficult to be determined. Usually the taxonomic characters are not discrete, and many characters are only expressed in adult but not in juvenile specimens. Therefore many questions are still unsettled in the identification ofthrough morphology. For example, the identification between the sister speciesandremains questionable (Chang, 1948; Hayashi, 1973; Imaoka., 1990; Liao and Clark, 1995; Liu., 2006).
Considering the morphological complexity, molecular phylogenetic analyses were carried out onliving in Chinese waters based on mitochondrial cytochrome oxidase subunit Ι (COΙ) sequence. Sequence data from GenBank for severalin the Sea of Japan were referred to for the analysis. The aim of this study is to resolve the general taxonomic issues, determine species boundaries and phylogenetic relationships within the genus.
2 Materials and Methods
2.1 Sample Collection
All Chinesetaxa were selected as samples except., due to the loss of type material of this species. A minimum of three specimens per taxon were sequenced. Specimens were collected from coastal waters of mainland China by the National Comprehensive Oceanography Survey in 1958–1960, Beibu Gulf (Gulf of Tonkin) Comprehensive Oceanographic Survey in 1959– 1962, China’s offshore scientific expedition (open and shared voyage) in 2009 and a scientific expedition to the East China Sea in 2010. All samples (vouchers) examined in this study are not type materials and listed in Table 1. Based on the collection data, the locations of sampling stations are illustrated in Fig.1. In addition, the specimens were photographed to provide a visual voucher for confirming the identification of the specimens used in this study (Fig.2). Finally, we also borrowed some specimens offrom the Department of Invertebrate Zoology, Smithsonian Institution, National Museum of Natural History (in Washington, D.C.) in order to check morphological characters (Table 2).
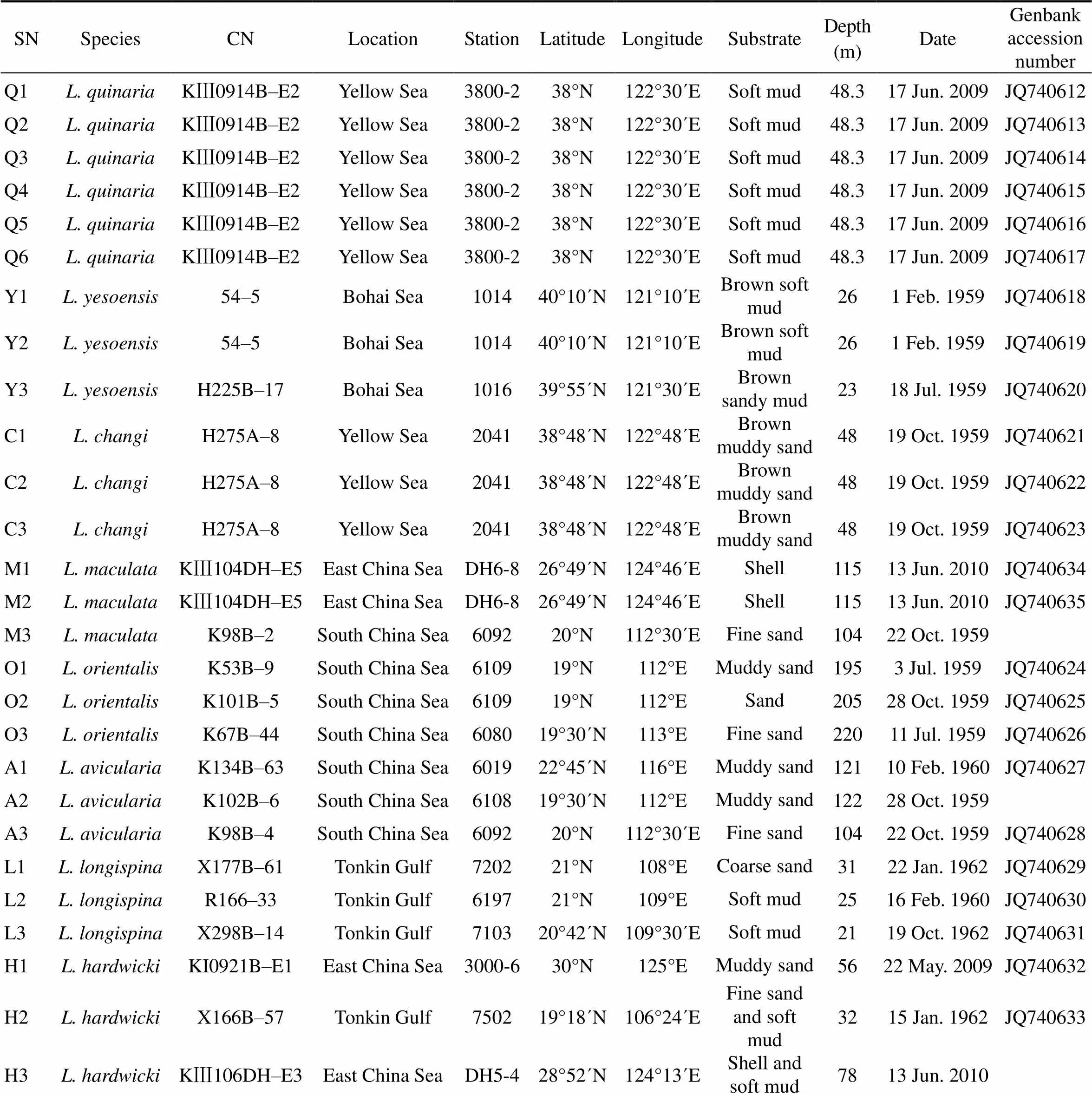
Table 1 Sampling information
Notes: SN, sampling number;CN, collection number.
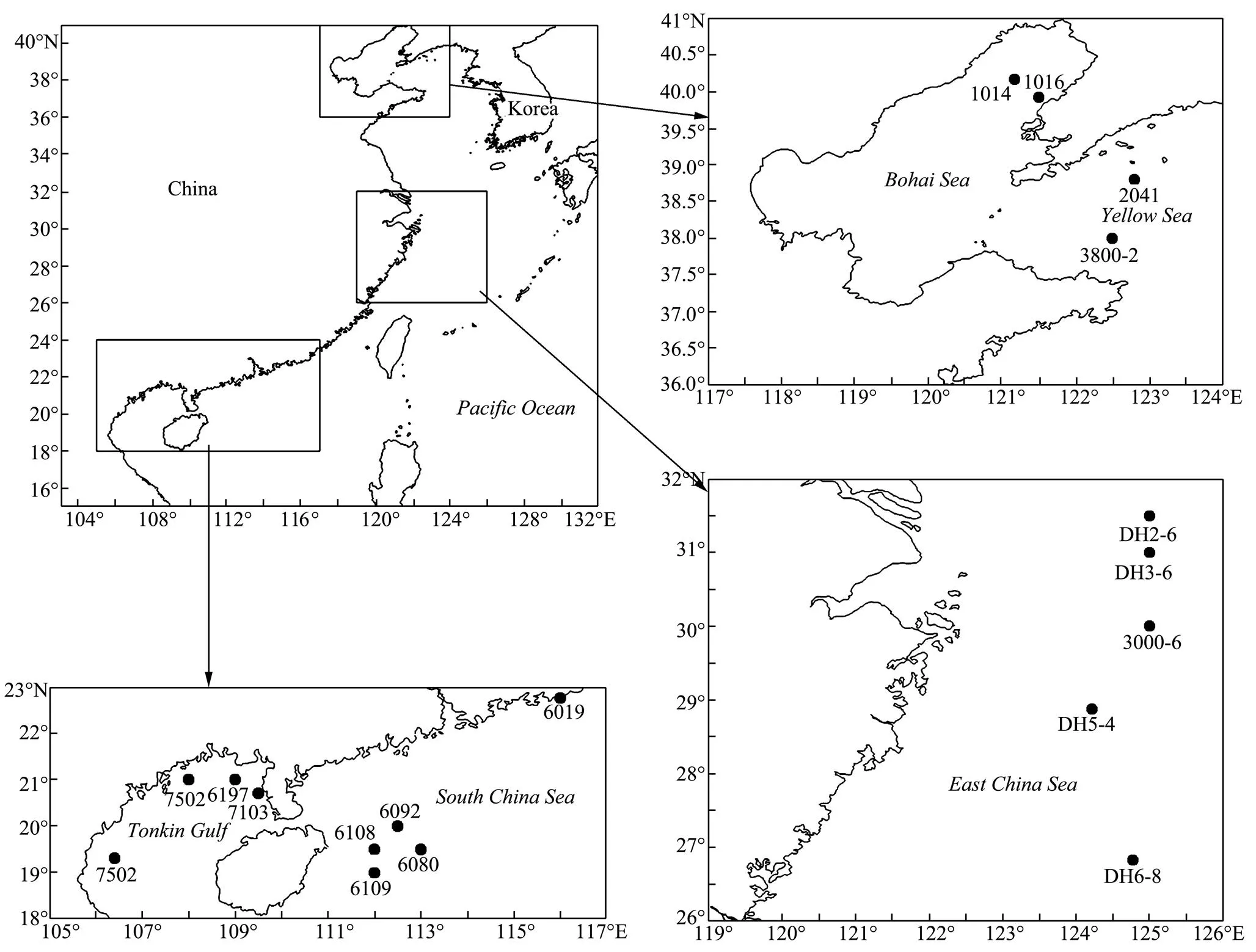
Fig.1 Locations of sampling stations.
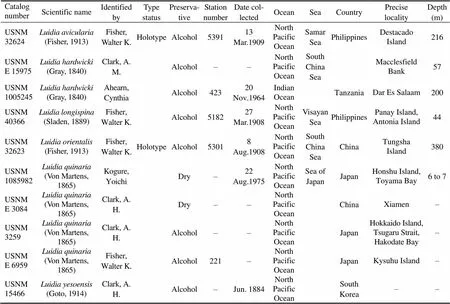
Table 2 Detailed information of the specimens in the NMNH Department of Invertebrate Zoology collections for the loan
Note: – not available.

Fig.2 Body photos of Luidia species and Craspidaster hesperus from Chinese waters. A,Luidia quinaria, Q2, abactinal view; B, Luidia quinaria, Q2, actinal view; C, Luidia quinaria, Q5, abactinal view, Note dark radiating bands on disk and arms; D, Luidia quinaria, Q5, actinal view; E, Luidia yesoensis, Y1, abactinal view; F, Luidia yesoensis, Y1,actinalview; G, Luidia changi, C1, abactinal view; H, Luidia changi, C1,actinal view; I, Luidia orientalis, O2,abactinal view; J, Luidia orientalis, O2,actinal view; K, Luidia avicularia, A3, abactinal view; L, Luidia avicularia, A3,actinal view; M, Luidia longispina, L2, abactinal view; N, Luidia longispina, L2,actinal view; O, Luidia hardwicki, H2,abactinal view; P, Luidia hardwicki, H2, actinalview; Q, Luidia maculata, M2, abactinal view; R, Luidia maculata, M2, actinalview; S, Craspidaster hesperus, W3,abactinal view; T, Craspidaster hesperus, W3,actinal view. Scale bar=10mm.
2.2 DNA Extraction, PCR Amplification and Sequencing
Genomic DNA was extracted from tube foot tissue using TIANamp Marine Animals DNA Kit (TIANGEN). A fragment of COI was amplified with standard polymerase chain reaction (PCR) and directly sequenced after purification. One pair of primers was used for PCR and sequencing: ECOΙA (5’ACCATGCAACTAAGACGAT- GA 3’) (Knott and Wray, 2000) and HCO (5’ TAAACTT- CAGGGTGACCAAAAATCA 3’) (Folmer., 1994). The amplification was conducted in a reaction mixture containing 0.125µL of exTaq polymerase (250U, TaKaRa), 2.5µL of exTaq buffer, 2.0µL of MgCl(25mmolL), 1.0µL of dNTPs (25nmol), 0.5µL of primers (10pmol), 5.0µL of template DNA and ddHO to a total volume of 25µL. Conditions for amplification included an initial denaturation at 95℃ for 5min followed by 30 cycles of 94℃ for 30s, annealing for 45s at 50℃, extension at 72℃ for 1min, which was followed by a final extension at 72℃ for 10min. PCR products were purified with a SanPrep Colum DNA Gel Extraction Kit (Sangon) and sequencing was conducted with ABI Big Dye protocols.
2.3 Sequence Alignment and Phylogenetic Analysis
The COΙ sequences alignments were performed using Clustal X with default parameters. Kimura 2-parameter genetic distances for COΙ were calculated using MEGA, Ver. 4 (Tamura., 2007). Maximum parsimony (MP), maximum likelihood (ML), neighbour joining (NJ) and Bayesian inference (BI) approaches were employed to construct phylogenetic trees. MP was implemented with the heuristic search option in PAUP 4.0b10 (Swofford, 2002). All informative bases in MP analyses were weighted equally and unordered. Bootstrapping proportions (BSP) (Felsenstein, 1985) with 1000 replicates were used for nodal evaluations. For ML, the evolutionary model that best fit the data set was selected using the likelihood ratio test (Goldman, 1993), and implemented in ModelTest version 3.06 (Posada and Crandall, 1998). Analyses were based on the selected model (TrN+G) using the heuristic search algorithm. An NJ phenogram was constructed and used as the initial topology for branch swapping. Bayesian inference analyses were performed with MrBayes 3.0 (Huelsenbeck and Ronquist, 2001). All Bayesian analyses were initiated with random starting trees. The site-specific rates were estimated during the run. Four Markov chains were used and the data set was run for 1×10generations to allow for adequate time of convergence. Trees were sampled every 100 generations. After approximately 20000 generations, the log-likely- hood values of each sampled tree stabilized. The last 18, 001 sampled trees were used to estimate the 50% majority rule consensus tree and the Bayesian posterior probabilities.
Twenty four samples were sequenced successfully and the sequences were used in the final analyses.(GenBank Accession Nos. AB183558 and NC006664) and(Grube, 1866) (GenBank Accession No. DQ380243) were included in these analyses and(Müller & Troschel, 1840) (Paxillosida: Astropectinidae) was selected as the outgroup.
3 Results
3.1 General Sequence Characteristics
The COI sequences consisted of 685 base pairs. In total, 202 nucleotide sites were potentially phylogenetic informative (29.5%). The average content of G/C was 43%.
3.2 Sequence Divergence
Kimura 2-Parameter distance is showed in Table 3. Genetic distances between individuals of the same taxa of Chinesetaxa ranged from 0.1% to1.3%, and those across the taxa ranged from 0% to 26.6%. Surprisingly, small genetic distances existed in the seven nominal species(,,,,,and), ranging from 0 to 1.6%. In contrast,individuals from the Sea of Japan and the Yellow Sea showed exceptional levels of genetic divergence (12.9%–13.1%).was highly distinct from othertaxa, with genetic distances ranging from 24.2% to 26.6%.
3.3 Tree Topology
Six unrooted cladograms were produced in the MP analyses, and the strict consensus tree included four main branches (Fig.3). Three clades were well supported by high bootstrap support (100%): Clade A (AB 183558+NC 006664), Clade B (a large clade with(Q1 to Q6),(Y1 to Y3),(C1 to C3),(O1 to O3),(A1 and A3),(L1 to L3) and(H1 and H2), and Clade C (M1+M2), which are also shown in the ML and BI trees with strong bootstrap support (see Figs.4 and 5). Furthermore, all three clades were well separated (13%–26%, see Table 4). By contrast, taxa within each clade exhibited small divergences (typically less than 1.0%, see Table 5). One basal node and some subordinate branches in the MP, ML and BI trees were not all well supported. Therefore, it was unable to resolve the relationship betweenDQ 380243 and othertaxa.
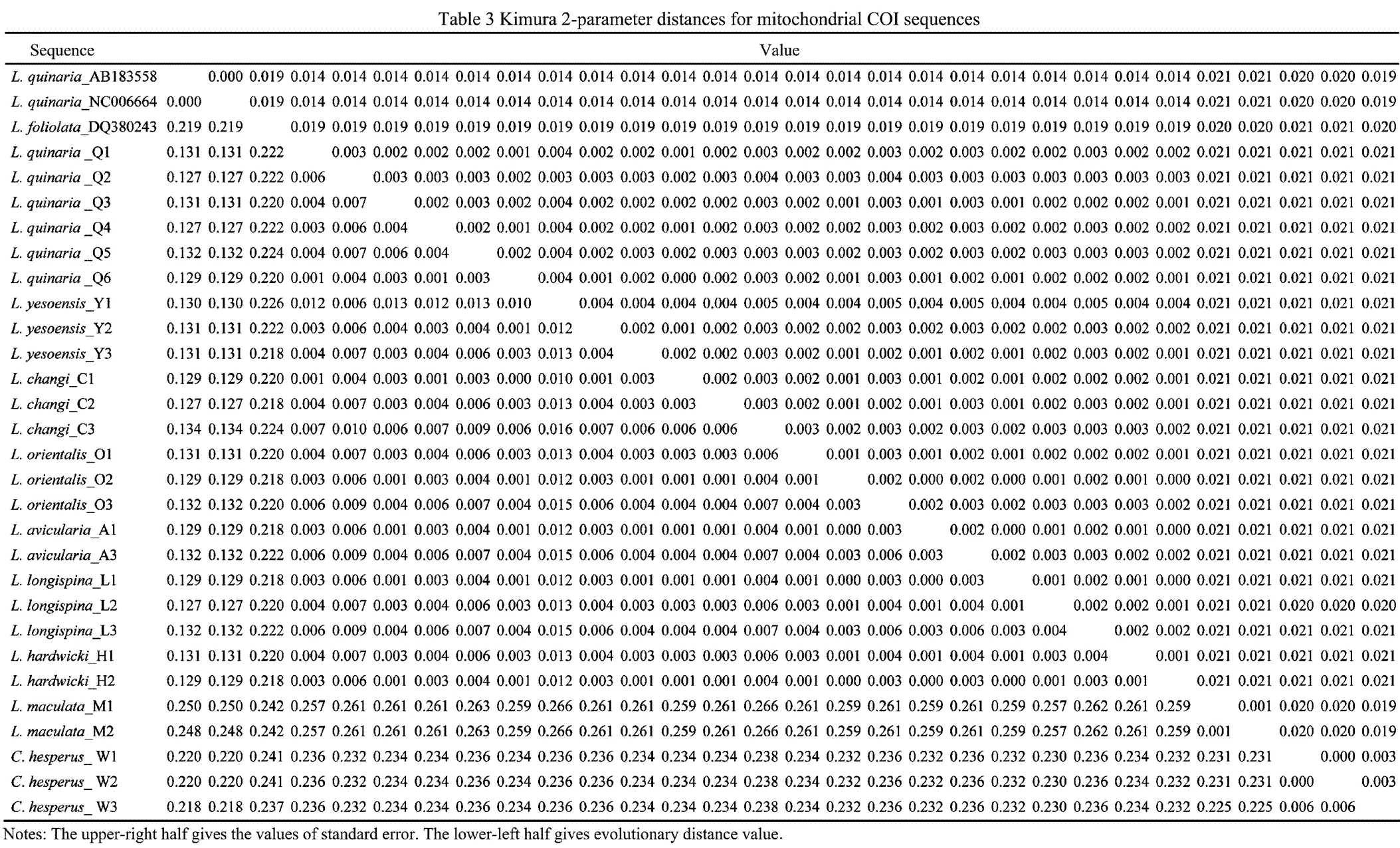
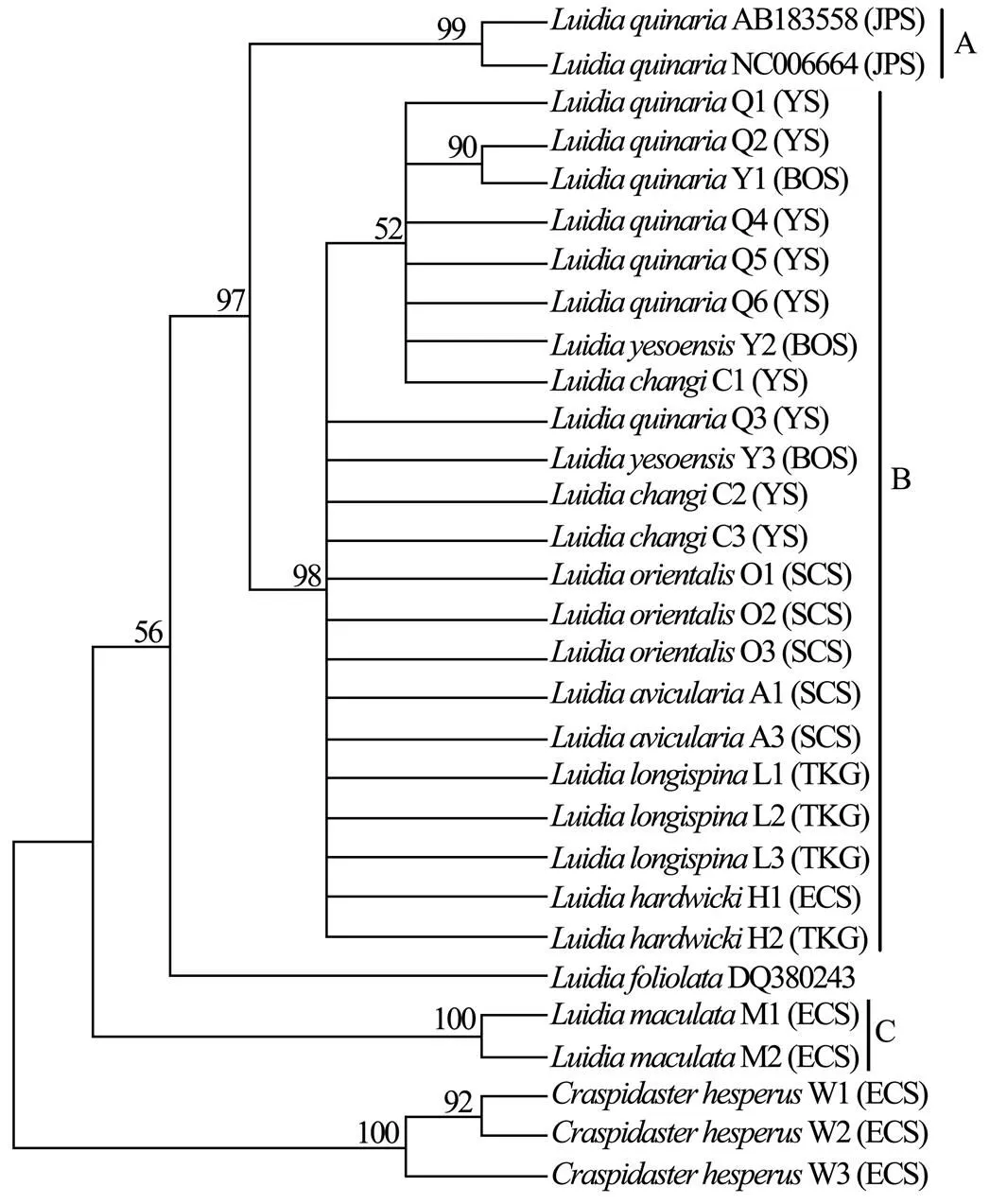
Fig.4 Maximum likelihood tree based on COΙ sequences. Bootstrap support values are indicated above nodes. See Table 1 for collection locality of specimens. (JPS, Sea of Japan; BOS, Bohai Sea; YS, Yellow Sea; ECS, East China Sea; SCS, South China Sea; TKG, Tonkin Gulf).
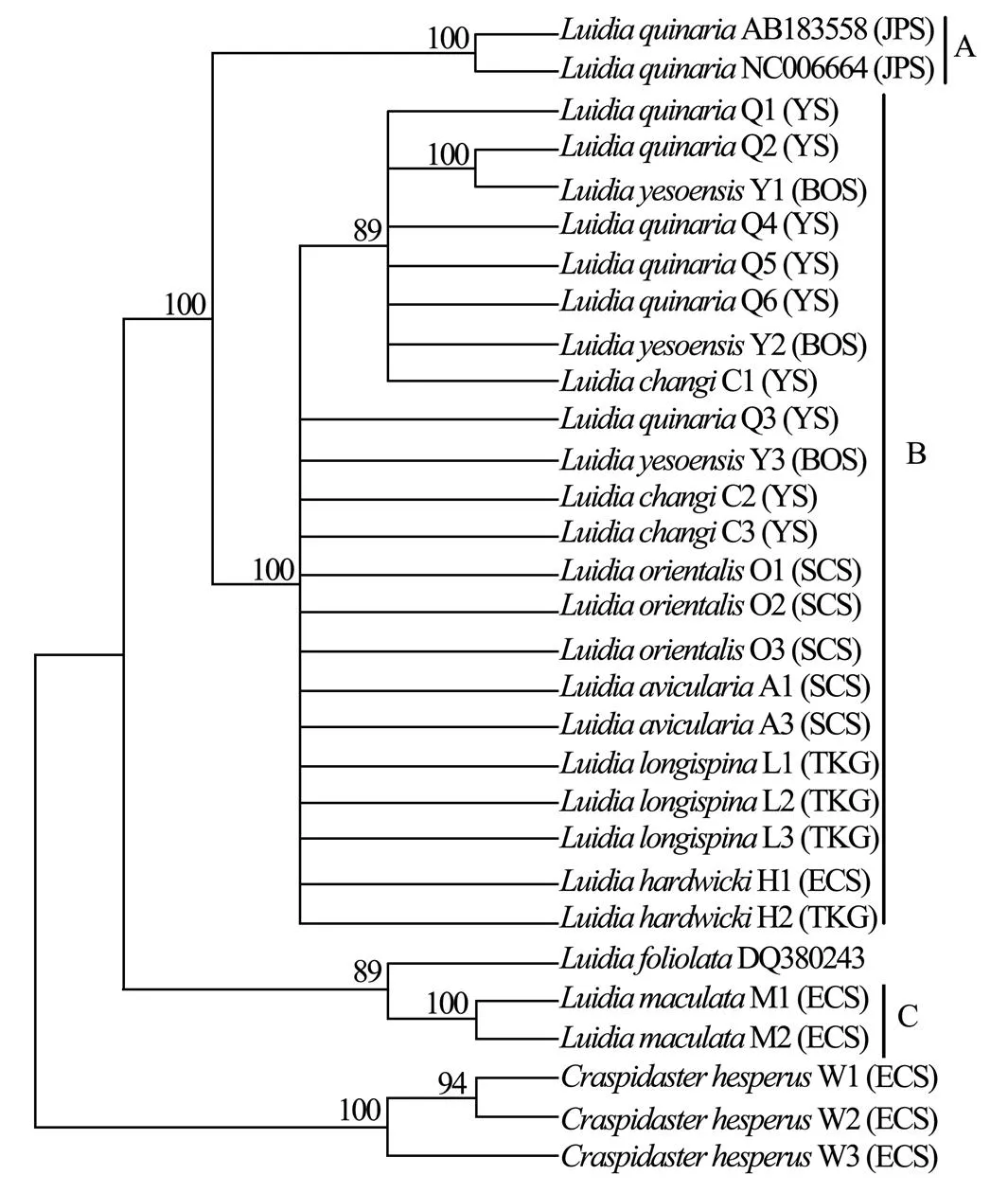
Fig.5 Bayesian inference tree based on COΙ sequences. Posterior probabilities are indicated above nodes. See Table 1 for collection locality of specimens. (JPS, Sea of Japan; BOS, Bohai Sea; YS, Yellow Sea; ECS, East China Sea; SCS, South China Sea; TKG, Tonkin Gulf).
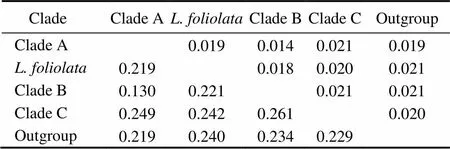
Table 4 Distances between clades
Notes: The upper-right half gives the values of standard error. The lower-left half gives evolutionary distance values.
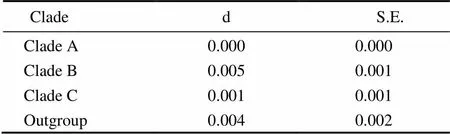
Table 5 Distances within clades
Notes: d, estimate; S. E., standard error.
4 Discussion
4.1 Molecular Phylogeny and the Classification of Döderlein (1920) in Chinese
Döderlein (1920) stated that there are four major groups inbased on morphological characters: Quinaria-Group, Ciliaris-Group, Clathrata-Group, and Alternata-Group. Systematic arrangement of some Chineseby Döderlein (1920) is shown in Table 6. In this study,,andare all clustered within the Clade B. Moreover,specimens are clustered into a separate clade as Döderlein suggested, not in the same group as the above three species. However,also appears in Clade B. Molecular data supportedas a member of the Quinaria-Group, which Döderlein considered belonging to the Ciliaris-Group.Döderlein (1920) did not assignto any group and neglected; our molecular data place these two species in the Quinaria- Group together with, which has been found and described after his classification. Though the number of taxa sampled is small and the sample is limited to Chinese species in the present study, the molecular phylogeny supports the distinction between the Quinaria-Group and Alternata-Group. With regard to the division of the Chinese Alternata/Quinaria Groups, the important characters are the presence or absence of pedicellariae close to the mouth and marked dark patches on the dorsal surface. From a broad perspective, further sampling oftaxa from other regions of the world would be highly desirable. It could have helped with the discussion or understanding of the Döderlein’s overall taxonomic groups.
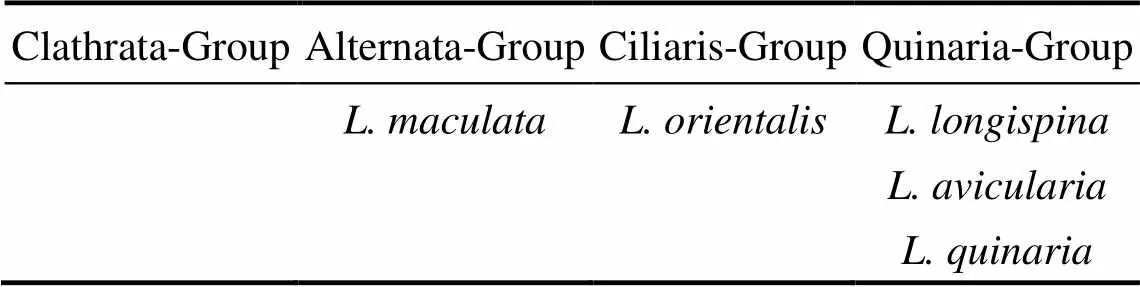
Table 6 Systematic arrangement of some Chinese Luidia by Döderlein (1920)
Note:indefinite.
4.2 Conflict Between Molecular Systematic and Traditional Taxonomy at the Species Level
Molecular systematic studiesoften conflict with traditional, morphology-based taxonomy, which is due tophenotypic difference arising from both environmental variation and genetic divergences. For example, taxonomically different species are sometimes genetically indistinguishable from each other (Williams, 2000). In contrast, a widespread ‘species’ is actually a species- complex, and cryptic speciation may have occurred (Zulliger and Lessios, 2010).
Our phylogenetic analyses revealed that the Chineseare divided into two genetically distinct monophyletic clades B and C. The first one includes seven nominal taxa,,,,,,and, which exhibited substantial morphological diversity (Fig.2A–P) and only show subtle differences for COI (ranging from 0 to 1.6%). In COI, the intraspecies divergence of Asteroidea ranges from 0 to 1.85% (Mean= 0.53%±0.13%) and the interspecies divergence of Asteroidea ranges from 2.17% to 22.85% (Mean=14.75%±0.62%) (Ward., 2008). The smallest interspecific distance was employed to define species boundaries in. Therefore, this low genetic divergence (less than 2% for COI) indicates that samples identified as,,,,,andvery likely belong to the same species. Considering that the incomplete speciation events can lead to conflicting genetic and morphological variation, it is possible that there is not enough time for COI to evolve diagnostic differences. As a result, many species share the identical DNA barcodes. However, the study of Ward. (2008)showed that DNA barcoding is likely to be an effective, accurate and useful method of species diagnosis for all five classes of Echinodermata. The present study also shows that genetic divergences ofallow species identification and reveal cryptic species within known taxa. For example, divergence between species ofand theother taxa,and theother taxa,and theother taxa exhibits favorable levels (>20%). Therefore, the possibility of many different species sharing identical DNA barcodes has been considered little. The second clade consists ofwhich is very common in southern China. It is the best-known Indo-Pacific species of, and can be easily recognizable by its large body, eight arms and dorsal surface patterned with dark and light colours, which do not disappear after preservation (Liao and Clark, 1995; Clark and Rowe, 1971). Based on our examination, the distinct diagnostic character exists in both preserved and fresh specimens. The validity ofis also supported by molecular results.
In the MP tree, theindividuals from the Sea of Japan form a separate clade with high support to otherindividuals from the Yellow Sea. Furthermore, the individuals from the Sea of Japan and the Yellow Sea show 12.9%–13.1% divergence for COI, which is substantially greater than typical intra-specific distance proposed by Ward(2008). Therefore, it is supposed there is a misidentification of the samples whose sequences are in GenBank, or a cryptic species exists. Recent molecular studies indicate that cryptic species are common in sea stars. For example, genetic data from COI and 16 allozyme loci suggest that there are two cryptic within the currently acceptedGray, 1840 (see Williams, 2000). Two mitochondrial gene regions and a nuclear gene region provide evidence of at least two biological species within the nominal species(Stimpson, 1862) (see Foltz., 2008). The result of molecule phylogeny ofbased on mtDNA sequences of 12S rRNA, 16S rRNA and COI revealed thatMüller & Tröschel, 1842 andDöderlein, 1888 are species-com- plexes; cryptic speciation might have occurred within each of these morphospecies (Zulliger and Lessios, 2010). The cryptic species derived from different areas suggests that ecological and physical conditions (., different vertical ranges or different water masses or geographical isolation) may prevent the flow of genes between the Yellow Sea and the Sea of Japan. The application of echinoderm COI divergence rates (3.5% per million years, Lessios., 2008) suggests that the individuals ofin the Yellow Sea and the Sea of Japan diverged before 1.85Ma. Although we were not able to obtain some samples (GenBank Accession Nos. AB183- 558 and NC 006664) for morphological re-examination, we obtained and examined another specimen (USNM 1085982) from the Japan Sea (Toyama Bay). Only minor morphological differences (mainly in the paxillar structure) can be found between specimens from the Sea of Japan and the Yellow Sea. The morphology is very similar, and current descriptions ofdo not permit delineation of species.
4.3 Reassessment of Current Diagnostic Morphological Characters
Given the subtle genetic divergences within seven Chinese taxa (,,,,,and), it would be more appropriate to consider them as one species. Therefore, our results suggest that several morphological characters need to be reassessed.
,,anddisplay considerable variations with regard to the pedicellariae on the adambulacral and inferomarginal plates in specimens from Chinese waters and those borrowed from the NMNH. Therefore, we doubt if their occurrence is sufficiently constant to serve as a character of specific weight. Moreover, the shape of abactinal paxillar spinelets (., central and peripheral spinelets being uniform or the central spinelets markedly longer than the rest on lateral paxillae) is only a minor morphological difference. The study of the sea starshows that the variation in spinelet morphology may be due to the action of waves (Xiao., 2011). We suggest that the shape of abactinal paxillar spinelets should not be considered as a character of specific weight. It is noteworthy that the number and form of large, erect inferomarginal spines are the major differences between,and, which may need to be re-evaluated.
In the northern waters of China, the identification of,andmight be confusing. Adults of(Fig.2C) are easily distinguished from the other species ofby their unique morphological characters, such as arms with a distinct dark area in disk center and along midradial line. However, young individuals (Fig.2A) are somewhat difficult to be distinguished from(Fig.2E) (abactinal surface uniformly dark grey or black, without radiating bands) since their arms are shorter and the dark radial midline is less distinct. Meanwhile,collected from the Yellow Sea differs fromandin details of abactinal plates, abactinal spinelets and body proportions (Liu., 2006). However, our molecular evidence reveals thatsamples from the Bohai Sea,samples andspecimens from the Yellow Sea might be conspecific. In other words, there may only be one species ofin the Yellow Sea and the Bohai Sea. Japanese samples from NMNH also reveal that the dark radial midline is variable in. Therefore we doubt whether the arm width and dark radial midline of upper side can be applied as characters with specific weight.
5 Conclusions
Phylogenetic analyses of Chinesebased on mitochondrial COI sequences support the distinction between Quinaria-Group and Alternata-Group as Döderlein (1920) assigned, but conflict with the current morphological taxonomy ofat the species level, except for. It is possible that the studied seven species,,,,,andare synonymized andGray, 1840 is the senior synonym. The results also show that cryptic speciation might have taken place inindividuals from Sea of Japan and the Yellow Sea. The present study shows that the combined analyses based on molecular gene sequence and morphological evidence represents a powerful technique for asteroid taxonomy.
Acknowledgements
This work was supported by the Knowledge Innovation Program of CAS (KSCX2-YW-N-0807), the Ministry of Science and Technology of the People’s Republic of China (2006FY110500), and IOCAS funding (2012IO 060104). The authors are grateful to Dr. Zhang Junlong at our laboratory (IOCAS) for his kind assistance with the figures. We thank David L. Pawson of Smithsonian Institution for facilitating the loan of some specimens of the genusfrom National Museum of Natural History. We also thank Dr. Song Linsheng (IOCAS) for his helpful comments.
Blake, D. B., 1973. Ossicle morphology of some recent asteroids and description of some west American fossil asteroids., 104: 1-59.
Chang, F. Y., 1948. Echinoderms of Tsingtao., 4 (2): 33-104.
Clark, A. M., and Downey, M. E., 1992.., 3. Natural History Museum Publications, London, 794pp.
Clark, A. M., and Rowe, F. W. E., 1971.. British Museum (Natural History), London, 238pp.
Clark, H. E. S., and McKnight, D. G., 2000. The marine fauna of New Zealand: Echinodermata: Asteroidea (sea-stars). Order Paxillosida & Notomyotida., 116: 1-196.
Djakonov, A. M., 1950. Starfish of the Soviet Union., 34: 1-203.
Döderlein, L., 1888. Echinodermen von Ceylon.,3: 822-846.
Döderlein, L., 1920. Die Asteriden der ‘Siboga’ Expedition. 2. Die Gattungand ihre Stammesgeschichte., 46b: 193-291, Figs.1–37.
Düben, M. W., and Koren, J., 1845. Untitled (in German; under name of Lóven)., 1: 436-440.
Fell, H. B., 1963. The phylogeny of sea-stars.., 246: 381-435.
Fisher, W. K., 1913. New starfishes from the Philippine Islands, Celebes and Moluccas., 46: 201-224.
Fleckenstein, J., 1985. Confidence limits on phylogenies: An approach using the bootstrap., 39 (4): 783-791.
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R., 1994. DNA primers for amplification of mitochondrial cytochromeoxidase subunit Ι from diverse metazoan invertebrates., 3: 294- 299.
Foltz, D. W., Nguyen, A. T., Kiger J. R., and Mah, C. L., 2008. Pleistocene speciation of sister taxa in a North Pacific clade of brooding sea stars (Leptasterias)., 154: 593-602.
Forbes, E., 1839. On the Asteriadae of the Irish Sea.,, 8: 114- 129.
Goldman, N., 1993. Statistical tests of models of DNA substitution., 36: 182-198.
Goto, S., 1914. A descriptive monograph of Japanese Asteroidea. 1. Archasteridae, Benthopectinidae, Porcellanasteridae, Astropectinidae, Luidiidae, Pentagonasteridae, Oreasteridae, Gymnasteriidae, Asterinidae.,, 29 (1): 1-808.
Gray, J. E., 1840. A Synopsis of the Genera and Species of the Class Hypostoma (,Linnaeus)., 6: 175-184, 275-290.
Grube, A. E., 1866. Einige neue Seesterne des heisigen zoologischen Museums., 43: 59-61.
Gui, L. P., Liu, C. Z., Sun, J. F., and Guo, Y. Q., 2011.Chemical Constituents fromGoto., 44 (6): 99-101.
Hayashi, R., 1973.,. Biology Laboratory, Imperial Household, Tokyo, 41-53.
Huelsenbeck, J. P., and Ronquist, F., 2001. MrBayes: Bayesian inference of phylogeny., 17: 754-755.
Imaoka, T., Irimura, S., Okutani, T., Oguro, C., Oji, T., Shigei, M., and Horikawa, H., 1990.. Vol. Ι. Japan Fisheries Resource Conservation Association Press, Tokyo, 159pp.
Knott, K. E., and Wray, G. A., 2000. Controversy and consensus in asteroid systematics: new insights to ordinal familial relationships., 40 (3): 382-392.
Lessios, H. A., 2008. The Great American Schism: divergence of marine organisms after the rise of the Central American Isthmus.,,,39:63-91.
Liao, Y. L., 2008. Echinodermata. In:. Liu, J. Y., ed., Science Press, Beijing, 845- 876.
Liao, Y. L., and Clark, A. M., 1995.. Science Press, Beijing, 614pp.
Liu, W., Liao Y. L., and Li, X. Z., 2006. A new sea-star species (Asteroidea: Luidiidae) from the South China Sea., 54 (2): 441-445.
Liu, W., Liao, Y. L., and Li, X. Z., 2006., a new sea star species (Echinodermata: Asteroidea: Luidiidae) from the Yellow Sea, with a review of two related species., 1315: 57-68.
Liu, W., Liao, Y. L., and Li, X. Z., 2007. Report on the sea-star species of Luidiidae (Echinodermata, Asteroidea) from the Chinese waters., 32 (1): 234-240.
Mah, C., and Hansson, H., 2012.Forbes, 1839. World Asteroidea database. Accessed through: World Asteroidea database at http://www.marinespecies.org/asteroidea/phia.hp? =taxdetails&id=123260 on 2012-02-16.
Martens, E. von., 1865. Ueber östasiatiche Echinodermen. 1. Japanische Seesterne. 2. Chinesische Seesterne., 31: 345-360.
Müller, J., and Troschel, F. H., 1840. Ueber die Gattungen der Asterien., 6: 318-326.
Müller, J., and Troschel, F. H., 1842.. Braunschweig, 134pp.
Posada, D., and Crandall, K. A., 1998. Modeltest: testing the model of DNA substitution., 14 (9): 817-818.
Schram, J. B., McClintock, J. B., Angus, R. A., and Lawrence, J. M., 2011. Regenerative capacity and biochemical composition of the sea star(Say) (Echinodermata: Asteroidea) under conditions of near-future ocean acidification., 407 (2): 266-274.
Sladen, W. P., 1889. Report on the Asteroidea collected by H. M. S. Challenger., 30: 1-935.
Sloan, N. A., 1980. Aspects of the feeding biology of asteroids., 18: 57- 124.
Stimpson, W., 1862. On new genera and species of starfishes of the family Pycnopodidae (Müll, and Trosch.), 8: 261- 273.
Swofford, D. L., 2002. PAUP*. Phylogenetic Analysis Using Parsimony and Other Methods. Version 4. Sinauer Associates, Sunderland, MA.
Tamura, K., Dudley, J., Nei, M., and Kumar, S., 2007. MEGA 4: molecular evolutionary genetics analysis (MEGA) software version 4.0.,24: 1596- 1599.
Ward, R. D., Holmes, B. H., and O’Hara, T. D., 2008. DNA Barcoding discriminates echinoderm species., 8: 1202-1211.
Williams, S. T., 2000. Species boundaries in the starfish genus., 136: 137-148.
Xiao, N., Liao, Y. L., and Liu, R. Y., 2011. Records of the genusGray, 1840 (Echinodermata: Asteroidea: Echinasteridae) from Chinese waters., 3115: 1-20.
Zulliger, D. E., and Lessios, H. A., 2010. Phylogenetic relationships in the genusGray (Paxillosida: Asteropectinidae) on a global scale: molecular evidence for morphological convergence, species-complexes and possible cryptic speciation., 2504: 1-19.
(Edited by Qiu Yantao)
10.1007/s11802-013-2158-0
ISSN 1672-5182, 2013 12 (3): 459-468
. Tel: 0086-532- E-mail: shazl@qdio.ac.cn
(September 14, 2012; revised October 9, 2012; accepted January 4, 2013)
© Ocean University of China, Science Press and Springer-Verlag Berlin Heidelberg 2013
杂志排行
Journal of Ocean University of China的其它文章
- Evaluation of Antitumor, Immunomodulatory and Free Radical Scavenging Effects of A New Herbal Prescription Seaweed Complex Preparation
- Shallow Water Body Data Processing Based on the Seismic Oceanography
- Prediction of China’s Submerged Coastal Areas by Sea Level Rise due to Climate Change
- A Homogeneous Linear Estimation Method for System Error in Data Assimilation
- The Suspended Sediment Concentration Distribution in the Bohai Sea, Yellow Sea and East China Sea
- Role of Ekman Transport Versus Ekman Pumping in Driving Summer Upwelling in the South China Sea
