Development of 101 Novel EST-Derived Single Nucleotide Polymorphism Markers for Zhikong Scallop (Chlamys farreri)
2013-07-29LIJiqinBAOZhenminLILingWANGXiaojianWANGShiandHUXiaoli
LI Jiqin, BAO Zhenmin, LI Ling, WANG Xiaojian, WANG Shi, and HU Xiaoli*
Development of 101 Novel EST-Derived Single Nucleotide Polymorphism Markers for Zhikong Scallop ()
LI Jiqin, BAO Zhenmin, LI Ling, WANG Xiaojian, WANG Shi, and HU Xiaoli
,,,,266003,
Zhikong scallop () is an important maricultured species in China. Many researches on this species, such as population genetics and QTL fine-mapping, need a large number of molecular markers. In this study, based on the expressed sequence tags (EST), a total of 300 putative single nucleotide polymorphisms (SNPs) were selected and validated using high resolution melting (HRM) technology with unlabeled probe. Of them, 101 (33.7%) were found to be polymorphic in 48 individuals from 4 populations. Further evaluation with 48 individuals from Qingdao population showed that all the polymorphic loci had two alleles with the minor allele frequency ranged from 0.046 to 0.500. The observed and expected heterozygosities ranged from 0.000 to 0.925 and from 0.089 to 0.505, respectively. Fifteen loci deviated significantly from Hardy-Weinberg equilibrium and significant linkage disequilibrate was detected in one pair of markers. BLASTx gave significant hits for 72 of the 101 polymorphic SNP- containing ESTs. Thirty four polymorphic SNP loci were predicted to be non-synonymous substitutions as they caused either the change of codons (33 SNPs) or pretermination of translation (1 SNP). The markers developed can be used for the population studies and genetic improvement on Zhikong scallop.
Zhikong scallop;; SNP; EST; HRM
1 Introduction
Zhikong scallop (Jones et Preston 1904) is an economically important maricultured species in China, and once accounted for 80% of the shellfish aquaculture production (Guo.,1999). With the rapid expansion of Zhikong scallop farming in recent 15 years, some problems, especially low productivity and diseases, have occurred mainly due to genetic degeneration. To effectively solve these problems, selective breeding programs for improving growth rates and disease resistance have recently been launched, which need a rich collection of molecular markers.
In the genomes of most organisms, single nucleotide polymorphisms (SNPs) are the most abundant and common variations, and have been widely used in high resolution genetic linkage map construction, QTL fine-mapping and population assessment (Williams., 2010). SNP markers developed from expressed sequence tags (ESTs) are particularly valuable, with which genes underlying quantitative traits are more easy to be found than markers from genome sequences (Muchero., 2011). By far, EST-derived SNPs have been reported in several marine mollusk species, such as Pacific oyster (Sau-vage., 2007), Pacific abalone (Qi., 2008, 2009), Mediterranean mussel (Vera., 2010), hard clam (Li., 2010), bay scallop (Li., 2009) and Yesso scallop (Liu., 2011). In Zhikong scallop, 44 EST-SNPs have been recently reported by Jiang. (2011). However, to carry out whole genome-based genetic surveys in molecular breeding programs, more molecular markers are appreciated.
With high-resolution melting (HRM) genotyping approach, 101 novel EST-based SNP markers were developed for Zhikong scallop from the putative SNPs found in its transcriptome dataset. We also annotated these polymorphic loci based on the transcriptome information. The markers developed should be useful for the population investigation and genetic improvement of Zhikong scallop.
2 Materials and Methods
EST data were generated by sequencing the transcriptome of Zhikong scallop using Roche 454 sequencing technology (unpublished). The raw reads were assembled into contigs and QualitySNP program was used to detect putative SNPs from contigs containing ≥4 reads. Single-base mutations represented by at least two reads in a contig were chosen for further analysis.
Each SNP site was genotyped with 2 PCR primers and one probe designed using Primer3 v4.0 (http://frodo.wi. mit.edu/primer3/). Primers designed must (1) be at least 20 bases in length, (2) amplify a fragment shorter than 150bp (preferablely around 100bp), (3) anneal to targets at temperatures ranging from 59℃ to 61℃, (4) contain 40%– 60% guanidine and cytosine, and (5) bound a single SNP, and the probe must (1) cover SNP in the middle, (2) be 20–35 bases in length, (3) anneal to the target at about 60℃, (4) be blocked at 3’ end by two random mismatched bases, and (5) overlap no bases with primers.
A total of 84 Zhikong scallop individuals were collected from 4 geographical populations in China, 36 from Changdao, Rongcheng and Rizhao, and 48 from Qingdao. Genomic DNA was extracted from the adductor muscle using standard phenol/chloroform method (Sambrook., 1989). Marker development was performed using the HRM assays discribed by Wang. (2009) with minor modifications. In detail, asymmetrical PCR was performed in a 10µL volume composing of about 15ng DNA, 0.1µmolLforward primer, 0.5µmolLreverse primer, 1.5mmolLMgCl, 2.0mmolLdNTP (each kind), 1U of rDNA polymerase (Takara) and 1×LCGreen Plus dye (Idaho Technology Inc.). The amplification was performed by denaturing at 95℃ for 5min; followed by 60 cycles of denaturing at 95℃ for 40s, annealing at 60℃ for 40s, and extending at 72℃ for 40s; and a final extension at 72℃ for 5min. In order to test the usefulness of primers, pooled genomic DNA from 48 individuals (12 for each of the four populations) was used as template and 5µL PCR product was separated on an 8% PAGE gel. Only those primer pairs amplifying expected products were used. The amplification was performed as described above, and then the corresponding probe was added to a final concentration of 2.0µmolL. After being denatured at 95℃ for 5min, the probe and PCR products were allowed to anneal at 25℃ for 30s. HRM analysis was performed on a LightScanner (Idaho Technology Inc.) by increasing temperature from 40℃ to 95℃ at a rate of 0.1℃swith optical signals collecting every 0.1℃. Data were retrieved and analyzed using the software of Light-Scanner.
To further evaluate these markers, 48 individuals from Qingdao were genotyped. Minor allele frequency and expected and observed heterozygosities (and) of these loci were estimated, and linkage equilibrium and Hardy-Weinberg equilibrium (HWE) were tested using Genepop v4.0.10 (http://genepop.curtin.edu.au/).
The ESTs containing polymorphic SNPs were annotated with BLASTx against nr and/or Swiss-Prot databases. A EST hits to either a known protein or a deposited protein if e-value less than 1e-5. The ORF in an EST was predicted using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf. html) in order to determine if different alleles of a SNP were synonymous.
3 Results and Discussion
In present study, 300 putative SNPs with conserved flanking sequences were identified from EST dataset and evaluated for their polymorphism and applicability in population genetic studies. Of the primer pairs designed to amplify the DNA fragments containing these SNPs, 96 (32.0%) were found to amplify fragments longer than expected. Introns may have lengthened the amplicons when genomic DNA was used as the template (Wang., 2008). As proved by cloning and sequencing, 87 of these primer pairs amplified fragments containing introns from 36bp to 1178bp in length.
For those species with reference genome sequences, amplification of intron containing fragments can be predicted and avoided; however, for species likewhose genome sequences are not available, shortening the size of amplicons in primer designing could be a strategy of avoiding intron-containing fragments. Another 28 (9.3%) primer pairs were discardedas they failed to get specific amplification products.
Of the remaining 176 primer pairs yielding products with expected sizes, 34 failed to generate decipherable probe binding curves. Probe binding to amplicons may be interfered either by undetected mutations, or nonspecific binding. Among 142 successfully genotyped loci, 41 were found to be monomorphic. Finally, 101 loci (33.7%) sur- vived polymorphism screening. Fig.1 showed an example of normalized melting curves for locus C42827S404_GA.
Further evaluation in the Qingdao population showed that, all the 101 loci were polymorphic with two alleles. The minor allele frequency ranged from 0.046 to 0.500. The observed and expected heterozygosities ranged from 0.000 to 0.925 and from 0.089 to 0.505, respectively. After Bonferroni correction for multiple tests, 15 loci were found to deviate significantly from HWE (Table 1). Significant linkage disequilibrium was detected between loci C49828S583_GT and C49739S111_GA.
With the advent of next generation sequencing technology, more and more ESTs are becoming available for a wide range of aquaculture species, from which molecular markers can be digged rapidly and effectively. For example, EST-derived SSRs have been developed for catfish (Liu., 1999), shrimp (Pérez., 2005), eastern oyster (Wang and Guo, 2007); EST-derived SNPs have been developed for Atlantic cod (Moen.,2008), turbot (Vera.,2011), Pacific oyster (Sauvage., 2007), hard clam (Li., 2010) and. From the transcriptome of Zhikong scallop we sequenced recently (data not shown), more than 20 thousands putative SNPs and more than 5 thousands microsatellites were discovered, which provided a large number of candidate loci for marker development.
For most non-model organisms, especially aquaculture species, current methods of SNP development are either low-throughput or expensive (Gravin., 2010) due to limited genetic and genomic information. With HRM technique, a SNP locus could be genotyped in 96 or 384 samples parallel in a few minutes (Li., 2012), and unlabeled probe HRM (Zhou., 2004) was reported to be more accurate and sensitive (Liew., 2007). In order to avoid extension of an unlabeled probe, its 3’ end was often blocked with C3 spacer, amino-modified C6, or phosphate (Dames., 2007). Instead of these ex-pensive traditional strategies, we blocked a probe at its 3’end with two mismatching bases.

Table 1 Summary of 101 SNP markers for Zhikong scallop (Chlamys farreri)
()
()

Locus IDPutativefunctionaPrimer sequence (5’-3’)Size (bp)HoHeMAFP-HWAmino acidb C34124S230_CGDynein heavy chain 10, axonemalF: GAGGGCCTCTTTAGACGAAT R: CTTCATCAGTGAAAGACCATGC Pb: CGGAGAATCTTcCCGAGGTGTGACT800.3400.379G 0.2500.469GGG-> GGC C34688S365_CTChitin deacetylase 3 F: ATCTCGCAGATTTCGAACCA R: GATGTTCCCGTCAGAACGAC Pb: TACATATCGCTcTCATCCCCTGCACA950.4720.401T 0.2740.298CTC-> CTT C34758S429_CTViral A-type inclusion protein F: AGCAGGACCTAGCCAAACTG R: CATGGTCCTGATCCTCCTGT Pb: CGGGACCAGAAcGTCACACTTCGGT1050.2410.214T 0.1201.000AAC-> AAT C34942S466_AGNAF: GTCATCAAATTGGCGGACTT R: CAGGGGCTAAATACGGTACG Pb: GCTGTTTGTGCGaTATCAAGCAAAGCA1060.4620.468G 0.3651.000ATA-> CTA Ile->Leu C35012S1133_ATKielin/chordin- like proteinF: TCGATTTCAGTACACGGAGAGA R: GCTGGGCAGTTATACGAACCPb: AGGTTGGAATGTGaGTGTGGCTGTAAT- CC750.4260.379A 0.2500.473GAG-> GTG Glu->Val C35668S542_AGCentrosomal protein of 164 kDaF: AGGGCTTGATGAGGTGATGT R: GCGTTGTCATGCTCATCTCT Pb: CATTCAGCAGCAaCAACAAAAGGAGAA940.2890.496A 0.4330.004CAA-> CAG C35935S329_GAPutative ferric- chelate reductase 1F: ACAAGCGCAGAGGGATACAG R: AGGGGTATCCCCCAATACAT Pb: GCCTGTGTTCCgTACCTCAGGATGT890.3460.289A 0.1730.324CGG-> TGG Arg->Trp C36105S330_TGWD repeat domain phosphoinositide- interacting protein 4F: TCATCACAGTTGACAGGAGACA R: ACAAGTGGACACACGGGAAC Pb: TCTGTATCTTTATTGtCCGTGTGTATAAG- GTAT1100.4910.464G 0.3580.768GTC-> GGC Val->Gly C36112S1135_CTPre-mRNA 3'- end-processing factor FIP1F: GGTGATCGGGACAGCTACAG R: TCCTTCTTGATCTGTGTTTTTCA Pb: CGTGAACGAGAcAGGTCAAGAGACG1040.6670.449T 0.3330.000†GAC-> GAT C36302S262_ATN-acylethanola- mine-hydroly- zing acid amidaseF: GGATTTCCCTCATGGACTTTG R: CCATGGTAACCTCTATTCAACACAPb: GAGGAAATAGTCaTGGTGATCGCAGTCT1100.3530.495A 0.4310.046----- C36849S1215_TAIntracellular protein transport proteinUSO1F: GAGGAAATTGAAGACAGGATGA R: TGGTGCTTCCTCTACAGCTTC Pb: AAAGTCAGTAAGTCtGCAGAAACTATTG- TACT1040.1890.203A 0.1130.502TCT-> TCA C37032S345_CTNAF: GTGGAATGTGCATCTTGGTG R: ATGAGTTGATGCTGGTTTCATC Pb: CGGAGCCCTAcGAGGGATACCTC1050.4530.426T 0.3020.749TAC-> TAT C37503S700_GACoatomer subunit gamma-2F: GCATCATGGGAGGAAGTAGG R: GCTTCTTCAAGAGTCTTCATGGT Pb: CAGAGAATGAACTgGAGGATACTTATGC- AG800.4810.500G 0.4520.784CTG-> CTA C37887S457_TAProtein Jade-3F: CTGACCAATGGAAGCAGGAT R: TTCACTGATGTTGGCAGTGG Pb: TGTCCAGGTACCtGTTAACTCCGAGCG830.6230.504A 0.4810.111CCT-> CCA C38102S339_TAE3 ubiquitin- protein ligase TRIM33F: GCCCCACAGCATTCAATAGT R: TCAGAAGATGCCAACGTTCA Pb: AAGCTGTCATGACAAtATTATTTGGAGT- GTTC1030.3020.374A 0.2450.257---- C39539S1242_CTSodium- and chloride-de- pendent glycine transporter 1F: ACACAACGCTGTCAAAGGTG R: CGTGGAGCAGACAGTGAAGA Pb: CGTTTACTGTGAcATTAGCGACACAACT1100.1510.500T 0.4530.000†GTC-> ATC Val->Ile C40293S161_TCPredicted protein F: GCTCAGACTTCGACATGATGC R: TTGCTGTCCGGTGGAATACTPb: GATGTGATAGTTCtGTGTCCTTTCCAGG- TA860.3920.342C 0.2160.418CTG-> CCG Leu->Pro
()
()

Locus IDPutative functionaPrimer sequence (5’-3’)Size (bp)HoHeMAFP-HWAmino acidb C41231S445_CACytosolic Fe-S cluster assembly factor narflF: ACATTAACAGATGAGCTGTGATAAA R: AACCATAGCATGGAATGAAGG Pb: TCAGTCTAAAAACTACAcATATTAAGAT- AAATCTCAC960.1510.490C 0.4150.000CAT-> AAT His->Asn C41331S764_GCHigh affinity choline transporter 1F: CCTTCCTCAGTTGATCTGTGC R: AACGTCCCAATGACGTATCC Pb: AGTGGACCAATGTgTACGGATCTGTAAG- C810.5000.500G 0.4541.000GTG-> GTC C41527S293_TCNAF: CCTGTTCCCGACACAGATTT R: CGATGACCTGGTGTTCTTTG Pb: GCGAAGTGTCAtGCAGTCTCCTACGC1060.3210.433C 0.3110.104CAT-> CAC C41740S1698_AGUbiquitin carboxyl-termi- nal hydrolase 2F: TCAAAGGTCGGCACGTCTAT R: CACACTCCTACAGGGTGACAAA Pb: CTGTCTGCATGGaCAAACTCAACTGTT740.1670.185G 0.1020.436GGA-> GGG C41766S952_AGNAF: TCATCTTGAAATGCATAATGGAG R: GAAACGGCTTGTTTCAAGGA Pb: TGGCGGTCACCaTACACCAACACACG890.4710.363A 0.2350.048TAT-> TAG C42372S299_ACNAF: TGCTGTAACATGTGACTCTGGA R: ATTACGGCAATCGTTTTGGA Pb: TCCAATCACACTATCaCATTTCAAAAGT- GTAAT1020.6850.491C 0.4170.004†---- C42827S404_GATranscriptional regulator ATRXF: ACACGGAACACAGACTGCAC R: GTGGCCTTGGAATCAATCTT Pb: CATCAAATATGATgACCCTGTTAGCTGC- TT940.3080.415A 0.2880.090GTC-> GTT C43109S299_GAProbable E3 ubiquitin-pro- tein ligase RNF217F: GACAGCCAGCTGTGGTATCTC R: ACCGCTACCACAAACAGAGG Pb: AGTGCCGTGATAgGTTGATATGCTGGT1070.0960.127A 0.0670.200CTA-> TTA C43423S605_AGNAF: CAGATGCAAACGATGAGAAGA R: TGTGGATTTTATCAGCAGCA Pb: CGCTAACCGTCTaATTGAAGACAACCTG900.6980.479A 0.3870.001†CTA-> CTG C45004S198_TCTrypsin alphaF: AAAGCAGAATTGTGGGAGGTT R: TGGCACCACAGATGTGACTT Pb: TGAATACCCAtGGCAGGCCTCGGA950.3850.478C 0.3850.237TGG-> CGG Trp->Arg C45270S783_TCAcetylcholine receptor subunit betaF: CGTCGCCTACCACTTTAATTTT R: CGGTAACATCAGCGAAATCC Pb: TGTGACGGTAGGAAtCCAGATATTAGAA- GAT980.4630.466T 0.3611.000ATT-> GTT Ile->Val C47045S426_GACoatomer subunit alphaF: TGCTGTTGGACTGAAGCTGA R: GATGGCCTCGGGGAATTT Pb: CTAGTACAGCAgTTACAGGTAGCCTA- GG880.4040.325A 0.2020.101CAG-> CAA C47562S784_AGArmadillo repeat-con- taining protein 4F: CCAGGAGCTCAAGGCAATAG R: ACTAGCACCTCCTCCGGTTG Pb: AGAGATGTTGGTaGCACTGCTGAATCT1040.1850.435G 0.3150.000†GTA-> GTG C48116S285_GACarboxylesterase 7F: CTTCCGTATTGGCCGAGAT R: TCCAGCGTCACGTGATATTC Pb: CCCAATAGGCGGaCTCTATCAACGGG890.5370.455G 0.3430.231GGA-> GGG C49262S732_CTDystroglycanF: CCAGGTTGAGTGGCAGAAAT R: CATGAAACAGCCGTCACTGT Pb: TCGTTCCAGAGcGCTCATACTCAG1100.0930.089T 0.0461.000GCT-> ACT Ala->Thr C49739S111_GAS-crystallin SL11F: GTCATGAACATGCATTGACAC R: AACGTCAATGTGAACCGAAC Pb: GCGTCACGCGgTTGATTGTCAGC650.3800.471A 0.3700.223---- C49828S583_GTDehydrogenase/ reductase SDR familymember 7F: CAGGGAGGACAGCCTTAGTT R: TTGAGGTTGACCAGCAGATG Pb: ATATTGGACCCAAgGTGTTGACTGAAAG- GT720.6420.505G 0.4910.057ACC-> ACA
()
()

Locus IDPutative functionaPrimer sequence (5’-3’)Size (bp)HoHeMAFP-HWAmino acidb C13455S296_AGCation transport regulator-like protein 2F: TTGCTGACTACGTCAGAGAACAC R: CGATGGAATTCCCAAATGTT Pb: ACCATCTATTTTCTTTaGATGAGACATTA- CGATTA1060.0570.157G 0.0850.001TTA-> TTG C13516S572_CTHypothetical protein PM8797T_ 01744 F: GATGTTCTTGGATGCCGTTT R: GATTTGACCAAGGCTTTCCA Pb: GTGAGGTAATGATGAAcAACAGCAGTAT- AATCAC770.0980.129T 0.0690.199GTT-> ATT Val->Ile C13520S236_CTYolk ferritinF: CACAGCGGCTAAAGAGGAAA R: TTCCAGATTGACGTTATCAAGG Pb: GGATTATCTGAAcATGCGAGGAGCATT970.5420.502T 0.4580.771AAC-> AAT C13588S690_CTProbable ATP- dependent RNA helicase DDX17F: CCTGGCAGTCCCACATTCTA R: TGTCAATATGTAAGGTAGTCTCTCTGG Pb: ATACACTTGGTCAGcTGTTTTCCAATGAA- TT1050.3920.472T 0.3730.239---- C13648S1364_CTPredicted protein F: ACTCCAGCGGAATTCTTCAA R: ACACCGTGCTTGTCCAGAA Pb: GAGACAGATCGcGCAGACACGCT960.3270.328T 0.2041.000CGC-> CGT C13664S764_CTZinc finger CCCH domain- containing protein 15F: TGGATTCTGTTTTGGAACATTG R: CAATGGAACAGTAATGCCACA Pb: AACTGTTGATGATGTcCTGAAACATTTAC- TGTA760.6800.492T 0.4200.007GGA-> GAA Gly-> Glu C13743S270_CTNAF: TGTCGAAAAATCGACACTAGAGAG R: GCAGCGCGTTTCTAGTCATT Pb: GTCAACAAACAGACcGAGAAAGTCATTT- TCCA820.2710.295T 0.1770.618TCG-> TCA C13864S561_GABaculoviral IAP repeat- containing protein 2F: AGTGAATCTGGCGTTTACGG R: CCACGTTCCACTGATGTGTC Pb: CGATGTCTGGTgCCTGTGGTGGACTT900.3000.285A 0.1701.000GCC-> ACC Ala->Thr C13871S591_TCUncharacterized transmembrane protein DDB_ G0289901F: TGCCATTTTCAACGACGATA R: CCGTTTCCTACTCCAACTCC Pb: TAATGGAGGCATtGTTGCTGGAGATGCT1080.2980.423C 0.2980.071ATT-> ATC C13981S547_GTNAF: AACCAACCAGTAACCTGACCA R: CTACCTCGGAAGTCGTTTCAA Pb: CAGTGGTCTTCgGAAACACTTCCGGAT1020.5290.505T 0.5000.784CCG-> CAG Pro->Gln C15003S544_CTNAF: GGACGTGTAGACCATGTATTCC R: ATCGCGACATTTTCCTTCAG Pb: ATTCGTTTTGTGAcGAGAGTCTAATGGA- AC840.2550.225T 0.1281.000GAC-> GAT C15117S465_TGSericin-2F: CGAAGGAGACCAAACTCTCG R: CAGGACGTCTTGGTAACTGATT Pb: CTAATGGACCTCtGGAGCAAACTGC1150.1520.142G 0.0761.000CTG-> CGG Leu->Arg C15256S166_GTNAF: GAGCAATAATCAGACTATGTTAAGACG R: TTAATGGAGACGTGGGTTCC Pb: CTCATGGAATAAGTgACCAGCGATTTTC- GT840.5710.505T 0.4900.401GTC-> GTA C15302S530_GCNAF: TGGGATATCCTCAACTTGAACC R: TGGCAGATATCCAGATATTCAAGA Pb: CGACCGTCTGTgACAAACTGGACTCA1070.5580.505C 0.4900.578GTC-> GTG C15320S180_GANAF: GCATACCTTATAACTGGTGATACATAA R: TGTCAACCTGAATAGGTCACATT Pb: CTTGATCAATGTGTCgATTTATAAGGTGA- ACAC1080.3700.388A 0.2590.732---- C15489S309_TAUncharacterized protein y4xOF: CTGGGCTTGCCGTATTAAAG R: ATGAGAGAAGCCGCTAGCAC Pb: AAAGTTGAAGTACTtGGAACAAAGTCCG- TGGA980.4340.505T 0.5000.404CTT-> CTA C15539S1133_CTSWI/SNF complex subunitSMARCC2F: ACTGGAGGCTCTGGAGATGT R: TGGGAAGTCGGAGAAAGTGT Pb: GTGGGTAGTCGcACACAAGATGAGAC850.3960.343T 0.2170.423CGC-> CGT
()
()

Locus IDPutative functionaPrimer sequence (5’-3’)Size (bp)HoHeMAFP-HWAmino acidb C15595S404_TANAF: TTGGAATTATGGTACTTGCATCA R: GGGAAGACATGATTAAGTTCAGG Pb: GTTTCATGAATATTTCTtATCAAGTTTAT- ACAATTGA950.6150.502A 0.4620.160TAT-> AAT Tyr->Asn C15628S1063_GA60S ribosomal export protein NMD3F: CCCAAACAGTTGGTGGAGTT R: TTGTTCGACACGGGAACAC Pb: GGTAGATTTTATAACgGAAAAGGAGCA- GAGTCT890.0770.110A 0.0580.142ACG->ACA C17329S372_ATGPutative ankyrin repeat protein RF_0381F: CGTGACAGCAACTCTTGAGC R: CCAGACACAGGATGAGTGGA Pb: TCGAACCATGCaGTACATAAGCGGGA870.1140.343A 0.2160.000†TGC-> AGC Cys->Ser C17433S1511_AGNAF: AGCCAAAACGGACTTCAGC R: CCAGCTGCAAATGTTCACAC Pb: CGCCGATAAGTCaGTCCCACTGTTCA1300.0000.498G 0.4360.000†TCA-> TCG C17452S1498_GADual specificity protein phosphatase CDC14AF: GCCACAAACGATCAACAACA R: GCCTTCAGAGGAGACATTGC Pb: CGGCCTATAGGaTCAAAGAGTGCTGT850.6300.505G 0.5000.101GGA-> GGG C17803S726_GANAF: CACAAGCAGCAGGAGAACAA R: AGTTTGCACGAAAACTGACACT Pb: CAGCCAATAGAACAgTCCGGTAATATAG- ACA730.5000.447A 0.3300.526CAG-> CAA C17942S612_TCNAF: CGTTGAACGAAGATGTATGCAG R: GGGTCAGCTTGCATTAGAGC Pb: GACCAGGATGTGGAtACAAATTTCTGTG- CAC730.5190.500T 0.4521.000GAT-> GAC C17986S114_TCNAF: TCGGAAGCCATACCTTTCAG R: ACGCAGTTGGAAATAGTGGA Pb: CTGACGTCCATCAGtGTTACTTACTATCC- TT1020.4230.498C 0.4420.402TGT-> CGT Cys-> Arg C18169S133_AGNAF: GGTGGAAATCCAGGGTCAG R: TGGAGGTTCCCCTGAAGATT Pb: AACTTCCCCTCaCCCGCCTGCGA770.1350.423G 0.2980.000†TCA-> TCG C18249S224_GTLP04489p [Drosophila melanogaster]F: TCTGCTCACATGACTACCTTCG R: TCGGATATTACCAGTGACTCTGC Pb: CGACGCAATGACgCAGTATGTAATCTA1050.2800.453G 0.3400.011ACG-> ACT C18475S582_AGNAF: TGACTTTCAATCTTACGTTTACGA R: AGATGAACTTGGAGCCGAGA Pb: ACACTTCCATCTCCaAGAACTTCTTCAA- CGA960.5000.447A 0.3300.524CTT-> CTC C18562S1428_GAIQ domain- containing protein DF: CAGGAACTGCAGCAATTGAA R: TTTCCTCTCAAATCCTGTTCG Pb: AGAATTTGATAACgGAGCACAGAGAAA- GGC800.2500.221A 0.1251.000ACG-> ACA C19634S751_GAAcid sphingo- myelinase-like phosphodiesterase 3bF: GACCTATCGCCAAACTCCAT R: TCGTCTTGGACACTGACGACPb: GAGAGAATGGGCgTGTCAAATAGTCCTC1080.7930.483A 0.3960.000†GTG-> ATG Val->Met C19848S175_TAHypothetical protein BRAF- LDRAFT_113741F: GCCAGGCTGGGATTATTTCT R: GGGTTTGTTTCTGACTTTGTTG Pb: CCTTAAGGAAGGtCAGGATCGTCGTAA810.2500.406A 0.2790.012GGA-> GGT C19996S325_TCChitotriosidase- 1F: GAGAAAACGAGACTTCGATGG R: GTTTATCTTCCGGCGGACTT Pb: CTCGACCTTGAtTGGGAGTACCCGCC790.4820.494C 0.4261.000GAT-> GAC C21017S1135_TAFK506-binding protein 14F: TGTTGTGACGTATAAAGGATGATG R: CGCTAAAACCGCCCTCTAGT Pb: TGTAACATGCGTTtAGTGTGTGTAGTGG- TT830.2080.383A 0.2550.002†TTA-> TAA Leu->†† C21312S837_GANAF: ATGGGGTCGATAATGTGTGC R: CCATTGGTCAAAGGTCGAGT Pb: GACTGTAACGATGgATACGTGACAGA- CTA1040.3700.369A 0.2411.000GGA-> GAA Gly->Glu
()
()
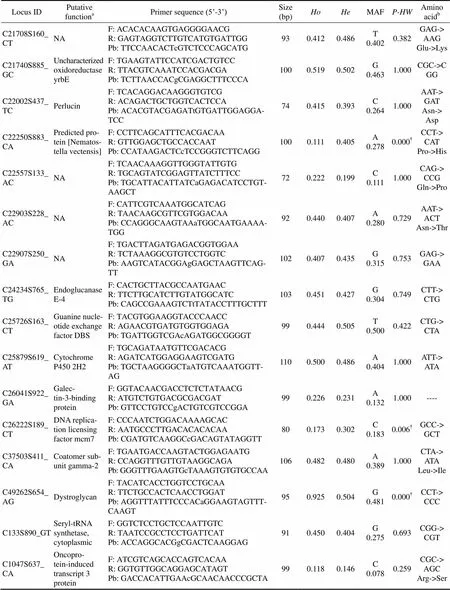
Locus IDPutative functionaPrimer sequence (5’-3’)Size (bp)HoHeMAFP-HWAmino acidb C21708S160_CTNAF: ACACACAAGTGAGGGGAACG R: GAGTAGGTCTTGTCATGTGATTGG Pb: TTCCAACACTcGTCTCCCAGCATG930.4120.486T 0.4020.382GAG-> AAG Glu->Lys C21740S885_GCUncharacterized oxidoreductase yrbEF: TGAAGTATTCCATCGACTGTCC R: TTACGTCAAATCCACGACGA Pb: TCTTAACCACgCGAGGCTTTCCCA1000.5190.502G 0.4631.000CGC->CGG C22002S437_TCPerlucinF: TCACAGGACAAGGGTGTCG R: ACAGACTGCTGGTCACTCCA Pb: ACACGTACGAGATtGTGATTGGAGGA- TCC740.4150.393C 0.2641.000AAT-> GAT Asn-> Asp C22250S883_CAPredicted protein [Nematostella vectensis]F: CCTTCAGCATTTCACGACAA R: GTTGGAGCTGCCACCAAT Pb: CCATAAGACTCcTCCGGGTCTTCAGG1000.1110.405A 0.2780.000†CCT-> CAT Pro->His C22557S133_ACNAF: TCAACAAAGGTTGGGTATTGTG R: TGCAGTATCGGAGTTATCTTTCC Pb: TGCATTACATTATCaGAGACATCCTGT- AAGCT720.2220.199C 0.1111.000CAG-> CCG Gln->Pro C22903S228_ACNAF: CATTCGTCAAATGGCATCAG R: TAACAAGCGTTCGTGGACAA Pb: CCAGGGCAAGTAAaTGGCAATGAAAA- TGG920.4400.407A 0.2800.729AAT-> ACT Asn->Thr C22907S250_GANAF: TGACTTAGATGAGACGGTGGAA R: TCTAAAGGCGTGTCCTGGTC Pb: AAGTCATACGGAgGAGCTAAGTTCAG- TT1020.4070.435G 0.3150.753GAG-> GAA C24234S765_TGEndoglucanase E-4F: CACTGCTTACGCCAATGAAC R: TTCTTGCATCTTGTATGGCATC Pb: CAGCCGAAAGTCTtTATACCTTTGCTTT1030.4510.427G 0.3040.749CTT-> CTG C25726S163_CTGuanine nucleotide exchange factor DBSF: TACGTGGAAGGTACCCAACC R: AGAACGTGATGTGGTGGAGA Pb: TGATTGGTCGAcAGATGGCGGGGT990.4440.505T 0.5000.422CTG-> CTA C25879S619_ATCytochrome P450 2H2F: TGCAGATAATGTTCGACACG R: AGATCATGGAGGAAGTCGATG Pb: TGCTAAGGGGCTaATGTCAAATGGTT- AG1100.5000.486A 0.4041.000ATT-> ATA C26041S922_GAGalectin-3-binding proteinF: GGTACAACGACCTCTCTATAACG R: ATGTCTGTGACGCGACGAT Pb: GTTCCTGTCCgACTGTCGTCCGGA990.2260.231A 0.1321.000---- C26222S189_CTDNA replication licensing factor mcm7F: CCCAATCTGGACAAAAGCAC R: AATGCCCTTGACACACACAA Pb: CGATGTCAAGGCcGACAGTATAGGTT800.1730.302C 0.1830.006†GCC-> GCT C37503S411_CACoatomer subunit gamma-2F: TGAATGACCAAGTACTGGAGAATG R: CCAGGTTTGTTGTAAGGCAGAPb: GGGTTTGAAGTGcTAAAGTGTGTGCCAA1060.4820.480A 0.3891.000CTA-> ATA Leu->Ile C49262S654_AGDystroglycanF: TACATCACCTGGTCCTGCAA R: TTCTGCCACTCAACCTGGAT Pb: AGGTTTATTTCCCACaGGAAGTAGTTT- CAAGT950.9250.504G 0.4810.000†CCT-> CCC C133S890_GTSeryl-tRNA synthetase, cytoplasmicF: GGTCTCCTGCTCCAATTGTC R: TAATCCGCCTCCTGATTCAT Pb: ACCAGGCACGgCGACTCAAGGAG910.4500.404G 0.2750.693CGG-> CGT C1047S637_ CAOncoprotein-induced transcript 3 proteinF: ATCGTCAGCACCAGTCACAA R: GGTGTTGGCAGGAGCATAGT Pb: GACCACATTGAAcGCAACAACCCGCTA990.1180.146C 0.0780.259CGC-> AGC Arg->Ser
Notes: For each probe sequence, SNP is indicated by a lower case letter. F: forward primer; R: reverse primer; Pb: probe; MAF: minor allele frequency;: observed heterozygosity;: expected heterozygosity;: exactvalue for Hardy-Weinberg equilibrium test;
: annotation information of the contig containing polymorphic SNP;: SNP in the codon of amino acid.: statistically significant after Bonferroni correction (<0.05); ---- indicated SNPs in non-coding regions;: stop codon.
Variations in genome or transcriptome, such as SNPs, could change codons or expression pattern. Of ESTs corresponding to the 101 polymorphic SNPs, 72 were found to encode important proteins such as intracellular protein transporter, ubiquitin-protein ligase, and transcriptional regulator (Table 1). Thirty one SNPs were located at the 1st two positions of codons and 2 at the 3rd position of codons. These SNPs were non-synonymous substitutions, which corresponded to different amino acid residues. They may change protein structure and function , or even the scallops’ phenotypes and traits. SNP at C21017S1135_TA was located at the 2nd position of leucine codon. One allele at this locus caused preterminated translation. Seven loci were located within non-coding regions. Another 60 loci were synonymous (Table 1).
In conclusion, we developed 101 EST-SNP markers for Zhikong scallop. These markers will be applicable for the population genetic studies and breeding of this species.

Fig.1 Normalized melting curves of locus C42827S404_GA.
The homozygous genotypes were either complementary with the probe (G/G), resulting in a melting peak at a high temperature, or completely not complementary with the probe (A/A), resulting in a melting peak at a low temperature; while heterozygous genotype (A/G) generates two melting peaks at both high and low temperatures, respectively.
Acknowledgements
Financial supports for this research were provided by the National Natural Science Foundation of China (No. 31130054), the National Basic Research Program of China (973 Program, 2010CB126406 and 2010CB126402), the National High-Tech R&D Program (863 Program, 2012 AA10A402 and 2012AA10A405), the National Key Technology R&D Program (2011BAD13B06), and the Earmarked Fund for Modern Agro-Industry Technology Research.
Choi, Y. S., Lee, K. S., and Park, D. H., 2005. Single nucleotide polymorphism (SNP) detection using microelectrode biochip array.,15 (10): 1938.
Dames, S., Margraf, R. L., Pattison, D. C., Wittwer, C. T., and Voelkerding, K. V., 2007. Characterization of aberrant melting peaks in unlabeled probe assays., 9 (3): 290-296.
Garvin, M. R., Saitoh, K., and Gharrett, A. J., 2010. Application of single nucleotide polymorphisms to non-model species: a technical review., 10: 915-934.
Guo, X., Ford, S. E., and Zhang, F., 1999. Molluscan auqaculture in China., 18 (1): 19-31.
Jiang, G. D., Li, J. Q., Li, L., Zhang, L. L., and Bao, Z. M., 2011. Development of 44 gene-based SNP markers in Zhikong scallop,., 3: 659-663.
Li, H., Zhu, D., Gao, X., Li, Y., Wang, J., and He, C., 2010. Mining single nucleotide polymorphisms from EST data of hard clam., 2: 69-72.
Liew, M., Seipp, M., Durtschi, J., Margraf, R. L., Dames, S., Erali, M., Voelkerding, K., and Wittwer, C., 2007. Closed- tube SNP genotyping without labeled probes/ a comparison between unlabeled probe and amplicon melting., 127: 341-348.
Liu, W. D., Li, H. J., Bao, X. B., He, C. B., Li, W. J., and Shan, Z. G., 2011. The first set of EST-derived single nucleotide polymorphism markers for Japanese scallop,., 42 (3): 456-461.
Liu, Z., Karsi, A., and Dunham, R. A., 1999. Development of polymorphic EST markers suitable for genetic linkage mapping of Catfish., 1 (5): 437-447.
Moen, T., Hayes, B., Nilsen, F., Delghandi, M., Fjalestad, K. T., Fevolden, S. E., Berg, P. R., and Lien, S., 2008. Identification and characterization of novel SNP markers in Atlantic cod: Evidence for directional selection., 9: 18.
Muchero, W., Ehlers, J. D., Close, T. J., and Roberts, P. A., 2011. Genic SNP markers and legume synteny reveal candidate genes underlying QTL forresistance and maturity in cowpea ((L) Walp)., 12: 8.
Pérez, F., Oritiz, J., Zhinaula, M., Gonzabay, C., Calderón, J., and Volckaert, F. A., 2005. Development of EST-SSR Markers by Data Mining in Three Species of Shrimp:,, and., 7 (5): 554-569.
Qi, H. G., Liu, X., and Zhang, G. F., 2008. Characterization of 12 single nucleotide polymorphisms (SNPs) in Pacific abalone., 8: 974-976.
Qi, H. G., Liu, X., Zhang, G. F., and Wu, F. C., 2009. Mining expressed sequences for single nucleotide polymorphisms in Pacific abalone., 40: 1661-1667.
Sambrook, J., Fritsch, E. F., and Maniatis, T., 1989.:. Cold Spring Harbor Laboratory Press, New York, 467-468.
Sauvage, C., Bierne, N., Lapègue, S., and Boudry, P., 2007. Single nucleotide polymorphisms and their relationship to codon usage bias in the Pacific oyster., 406: 13-22.
Syvanen, A. C., 2001. Accessing genetic variation: genotyping single nucleotide polymorphisms., 2 (12): 930-942.
Vera, M., Alvarez-Dios, J. A., Millan, A., Pardo, B. G., Bouza, C., Hermida, M., Fernandez, C., Herran, R. D. L., Molina- Luzon, M. J., and Martinez, P., 2011. Validation of single nucleotide polymorphism (SNP) markers from an immune expressed sequence tag (EST) turbot,, database., 313: 31-41.
Vera, M., Pardo, B. G., Pino-Querido, A., Alvarez-Dios, J. A., Fuentes, J., and Martinez, P., 2010. Characterization of single-nucleotide polymorphism markers in the Mediterranean mussel,., 41: 568-575.
Wang, S., Sha, Z., Sonstegard, T., Liu, H., Xu, P., Somridhivei, B., Peratman, E., Kucuktas, H., and Liu, Z., 2008. Quality assessment parameters for EST-derived SNPs from catfish., 9: 450.
Wang, S., Zhang, L. L., Meyer, E., and Matz, M. V., 2009. Construction of a high-resolution genetic linkage map and comparative genome analysis for a reef-building coral., 10: R126.
Wang, Y., and Guo, X., 2007. Development and characterization of EST-SNP markers in the Eastern oyster., 9 (4): 500-511.
Williams, L. M., Ma, X., Boyko, A. R., Butamante, C. D., and Oleksiak, M. F., 2010. SNP identification, verification, and utility for population genetics in a non-model genus., 11: 32.
Zhou, L., Myers, A. N., Vandersteen, J. G., Wang, L., and Wittwer, C. T., 2004. Closed-tube genotyping with unlabeled oligonucleotide probes and a saturating DNA dye., 50 (8): 1328-1335.
(Edited by Qiu Yantao)
10.1007/s11802-013-2007-1
ISSN 1672-5182, 2013 12 (3): 403-412
. Tel: 0086-532-82031970 E-mail: hxl707@ouc.edu.cn
(April 11, 2012; revised June 7, 2012; accepted July 18, 2012)
© Ocean University of China, Science Press and Springer-Verlag Berlin Heidelberg 2013
杂志排行
Journal of Ocean University of China的其它文章
- Evaluation of Antitumor, Immunomodulatory and Free Radical Scavenging Effects of A New Herbal Prescription Seaweed Complex Preparation
- Shallow Water Body Data Processing Based on the Seismic Oceanography
- Prediction of China’s Submerged Coastal Areas by Sea Level Rise due to Climate Change
- A Homogeneous Linear Estimation Method for System Error in Data Assimilation
- The Suspended Sediment Concentration Distribution in the Bohai Sea, Yellow Sea and East China Sea
- Role of Ekman Transport Versus Ekman Pumping in Driving Summer Upwelling in the South China Sea
