Lack of association between genetic polymorphisms in cytokine genes and tumor recurrence in patients with hepatocellular carcinoma undergoing transplantation
2013-05-22
Hangzhou, China
Lack of association between genetic polymorphisms in cytokine genes and tumor recurrence in patients with hepatocellular carcinoma undergoing transplantation
Li-Ming Wu, Lin Zhou, Jun Xu, Ba-Jin Wei, Jun Cheng, Xiao Xu, Bin Xi, Hai-Yang Xie and Shu-Sen Zheng
Hangzhou, China
BACKGROUND:Recurrence of hepatocellular carcinoma (HCC) after liver transplantation (LT) remains one of the most common causes of poor long-term survival. However, the host genetic factors affecting increased risk of tumor recurrence after transplantation have not been thoroughly elucidated. The present study was designed to investigate the association of cytokine gene polymorphisms with the risk of tumor recurrence in LT patients for HCC.
METHODS:Eleven single-nucleotide polymorphisms within the promoter regions of 7 cytokine genes, i.e., the IL-1 family (IL-1α and IL-1β), IL-6, IL-8, IL-10, TNF-α, and TGF-β1, were genotyped in 93 HCC patients treated with LT using DNA sequencing. The association between these polymorphisms and the risk of tumor recurrence was evaluated while controlling confounding clinical variables.
RESULTS:The genotype frequency of IL-10 -1082 A/G in patients with and without recurrence of HCC was AA 83.3%, GA 16.7% and AA 97.6%, GA 2.4%, respectively. The association between IL-10 -1082 GA and recurrence was signif i cant (P=0.033). No other single-nucleotide polymorphism in the cytokine gene was found to be associated with recurrence. Kaplan-Meier survival curves showed that the homozygous AA patients had a signif i cantly longer mean recurrence-free survival thanheterozygous GA patients (23.5 vs 5.7 months,P=0.001). However, multivariate analysis failed to reveal that the GA genotype of IL-10 -1082 A/G was an independent indicator of recurrence.
CONCLUSIONS:This study suggests the lack of association of selected cytokine gene polymorphisms with HCC recurrence after LT in the Han Chinese population. The fi nding does not exclude the idea that other cytokine polymorphisms could act as candidate biomarkers of disease prognosis.
(Hepatobiliary Pancreat Dis Int 2013;12:54-59)
hepatocellular carcinoma;cytokine; polymorphism; recurrence;
Introduction
Primary liver cancer in men is the fi fth most common cancer and the second most common cause of cancer-related deaths worldwide. In women, it is the seventh most frequent cancer and the sixth leading cause of cancer death. And it consists mainly of hepatocellular carcinoma (HCC).[1,2]Liver transplantation (LT) has been considered as a potentially curative option for HCC patients. However, the recurrence of HCC in patients undergoing LT remains an adverse factor for post-LT survival. Currently there is still lacking biomarkers, especially pre-transplant markers, to reliably predict tumor recurrence after LT in spite of extensive clinical and basic research. Over the last few years, many investigators have searched for reliable genetic markers in an attempt to better distinguish subtypes of patients with different risk of HCC recurrence after LT. We have retrospectivelyanalyzed some genetic variants on HCC recurrence after LT, based on the clinical database from the cohort of HCC patients who underwent LT at our institution.[3-5]However, nothing is known about the effects of genetic variants of the cytokine gene on tumor recurrence of LT recipients for HCC.
Cytokines take part in the construction of a highly complex and coordinated network where they modulate their own synthesis and that of other cytokines and cytokine receptors.[6]Numerous cytokines could be produced by immune cells in response to both antigenspecif i c and nonspecif i c stimuli. This process is critical to the outcome of immune-mediated disease, including cancer. HCC is also considered a cytokine-responsive tumor.[7]The clinical course of HCC patients may thus be associated with the capacity of producing distinct cytokines.
In the past few years, efforts have focused on the detection of single-nucleotide polymorphisms (SNPs) within cytokine gene sequences, particularly within the promoter regions. And several polymorphisms may be associated with different levels of gene transcription. Therefore, genetic studies[8-19]have attempted to correlate these cytokine polymorphisms with human cancers. Studies[9-13]have shown that some of the identif i ed loci in cytokine genes, including IL-1β, TNF-α, and TGF-β1, seem to contribute to HCC susceptibility. But much conf l ict and uncertainty in the literature with regard to cancers, genes, and SNPs need to be identif i ed. To evaluate the role of cytokines in the immunologic events after LT, we have investigated selected SNPs in cytokine genes and their association with acute rejection and recurrence of hepatitis B in 186 Chinese LT recipients.[20]Despite these advances, the alleles contributing to the genetic risk to HCC recurrence after transplantation remain undiscovered.
Based on the potential functional relevance of cytokines in the progress of HCC and LT, we hypothesize that the functional SNP in cytokine promoter may contribute to HCC recurrence after LT. In the present study, which included ethnically homogeneous Chinese patients, the effect of 11 SNPs in cytokine genes including IL-1 (IL-1α -889 C/T and IL-1β -31 T/C, IL-1β -511 C/T), IL-6 -572 G/C, IL-8 -251 T/A, IL-10 (-1082 A/G, -819 T/C, -592 A/C), TNF-α -308 G/A and TGF-β1 (-509 T/C, +869 C/T) was examined among patients with or without recurrence of HCC after LT.
Methods
Study population
The study was conducted on blood specimens from 93 HCC patients who had undergone LT between February 2003 and February 2006 at our hospital. The patients comprised Han Chinese only. The number of patients in this study was the same as that reported previously.[4]For immunosuppression, the regimen consisted of triple drugs of cyclosporine (CsA) or tacrolimus (FK506), mycophenolate (MMF) and prednisone. The treatment of patients with positive HBV DNA has been described in our previous study.[20]Blood specimens were obtained before the cirrhotic liver was removed. Histopathologic examination conf i rmed the diagnosis of the patients. Clinicopathologic variables including portal vein tumor thrombi (PVTT), preoperative alpha-fetoprotein (AFP) level, histopathologic grading, tumor size, and tumor number were available for this sample set. AFP test, ultrasonography, chest X-ray, and emission computed tomography were performed every 3 months for the fi rst 2 years and semiannually thereafter to monitor tumor recurrence or metastasis. Only patients with imaging evidence, either intrahepatically or extrahepatically (lymph nodes, distant metastases) were considered to have tumor recurrence. And the patient with an increase of AFP but without radiologic evidence of a new tumor was excluded. This study was approved by the Ethical Review Committee of the hospital. All the 93 patients were followed up from the date of operation until death. The time of follow-up was described previously.[3]
Genotyping
Genomic DNA was isolated from EDTA-anticoagulated whole blood by using QIAamp DNA blood mini kit (QIAGEN GmbH, Hilden, Germany). All SNPs of the cytokine gene were genotyped in our sample set by PCR direct sequencing. PCR and sequencing primers were listed in Table 1. The conditions of PCR amplif i cation and the mass extension reaction were available on request. Genotyping was carried out in a blind way in which the status of the patients was unknown to the performers. To assess the reliability of genotyping, we randomly chose more than 10% of the samples to sequence twice and all results showed 100% concordance. We randomly sequenced more than 10% of the samples twice to assess the reliability of genotyping.
Statistical analysis
Data analysis was performed using the SPSS statistical software version 16.0 for Windows (SPSS Inc., Chicago, IL., USA). The observed value of each genotype was compared to the expected value by the Hardy-Weinberg equilibrium. For statistical comparisons, either the Chi-square test or two-sided Fisher's exact test was used appropriately. Recurrence-free survival (RFS)was def i ned as the interval from the date of operation to the date of documented tumor recurrence or the last follow-up visit if recurrence did not occur. RFS curves were calculated with the Kaplan-Meier method and compared with the log-rank test. Then the association between the positive cytokine gene SNPs, clinical characteristics of patients and RFS was further analyzed by univariate Cox regression analysis. Variables identif i ed in the univariate analysis as signif i cant were used in multivariate analysis using Cox's proportional hazard model. All tests were two tailed and aPvalue less than 0.05 was considered statistically signif i cant.

Table 1.Primers for PCR and sequencing of cytokine genes
Results
In the 93 HCC patients, 85 (91.4%) were male and 8 (8.6%) female. Their median age was 47 years (19-67). Eighty-f i ve patients (91.4%) were infected with HBV, and one of them had double infection with HBV andHCV. The clinical characteristics of patients were derived from our previous study.[3]The median followup time was 10.7 months (1-45.9).

Table 2.Genotypes frequencies of cytokine genes in patients according to the absence or presence recurrent HCC after LT
The genotyping results are shown in Table 2. Because of a genotyping error of some samples, the number of samples detected for each polymorphism was not exactlyequal. To detect the association between genotype and tumor recurrence, we analyzed the genotype distribution of all SNPs in the patients. The genotype frequency of the IL-10 -1082 A/G in patients with and without recurrence of HCC was AA 83.3%, GA 16.7% and AA 97.6%, GA 2.4%, respectively (Table 2). The G allele was obviously rich in patients with recurrence compared with those without recurrence (8.3% vs 1.2%). Allele frequencies of the two subgroups were both in the Hardye-Weinberg equilibrium (P>0.05, data not shown). The association was signif i cant between IL-10 -1082 GA and recurrence (P=0.033). But no other SNP in the cytokine gene was associated with recurrence.
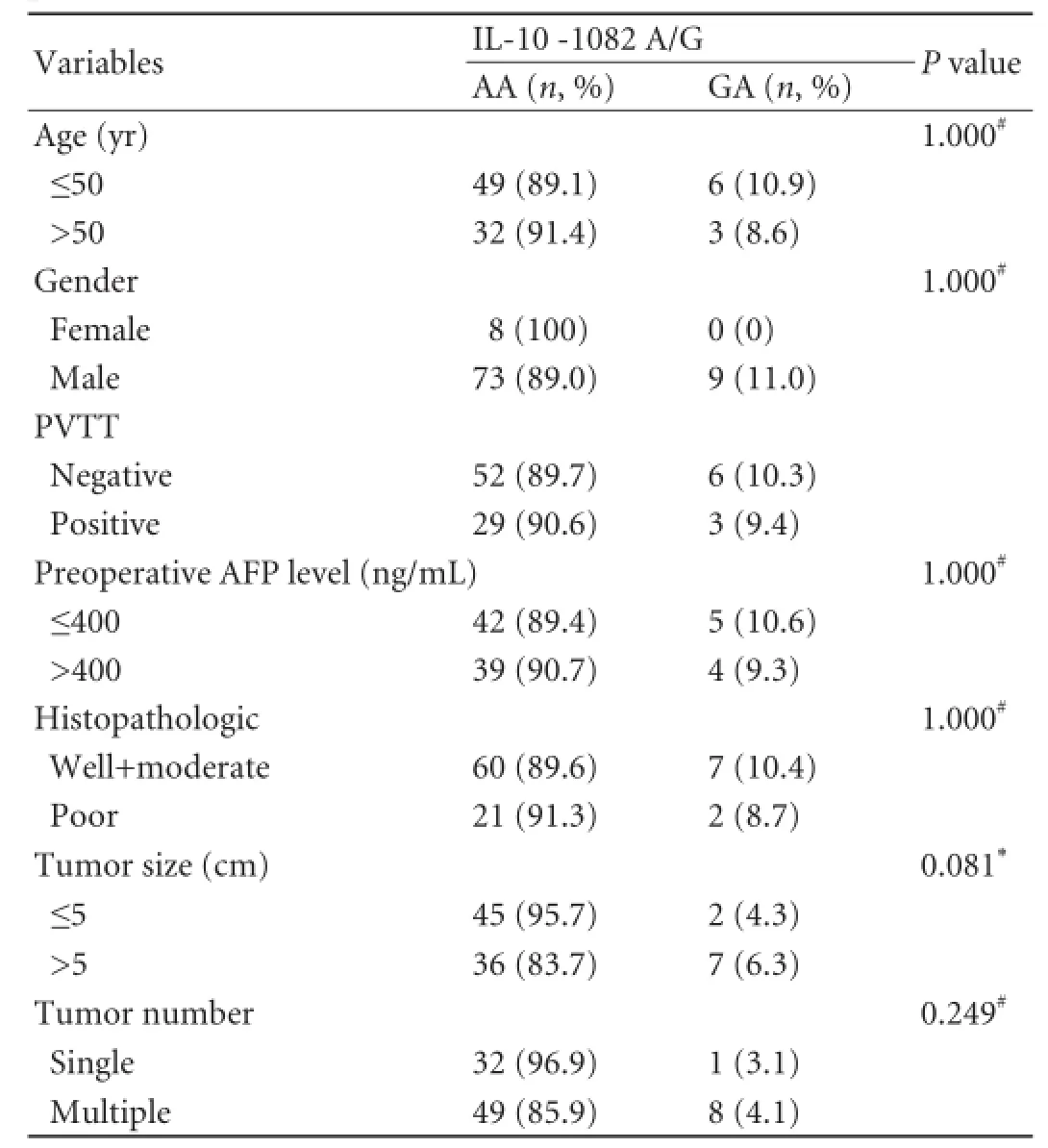
Table 3.Association of IL-10 -1082 genotypes with clinicopathological parameters
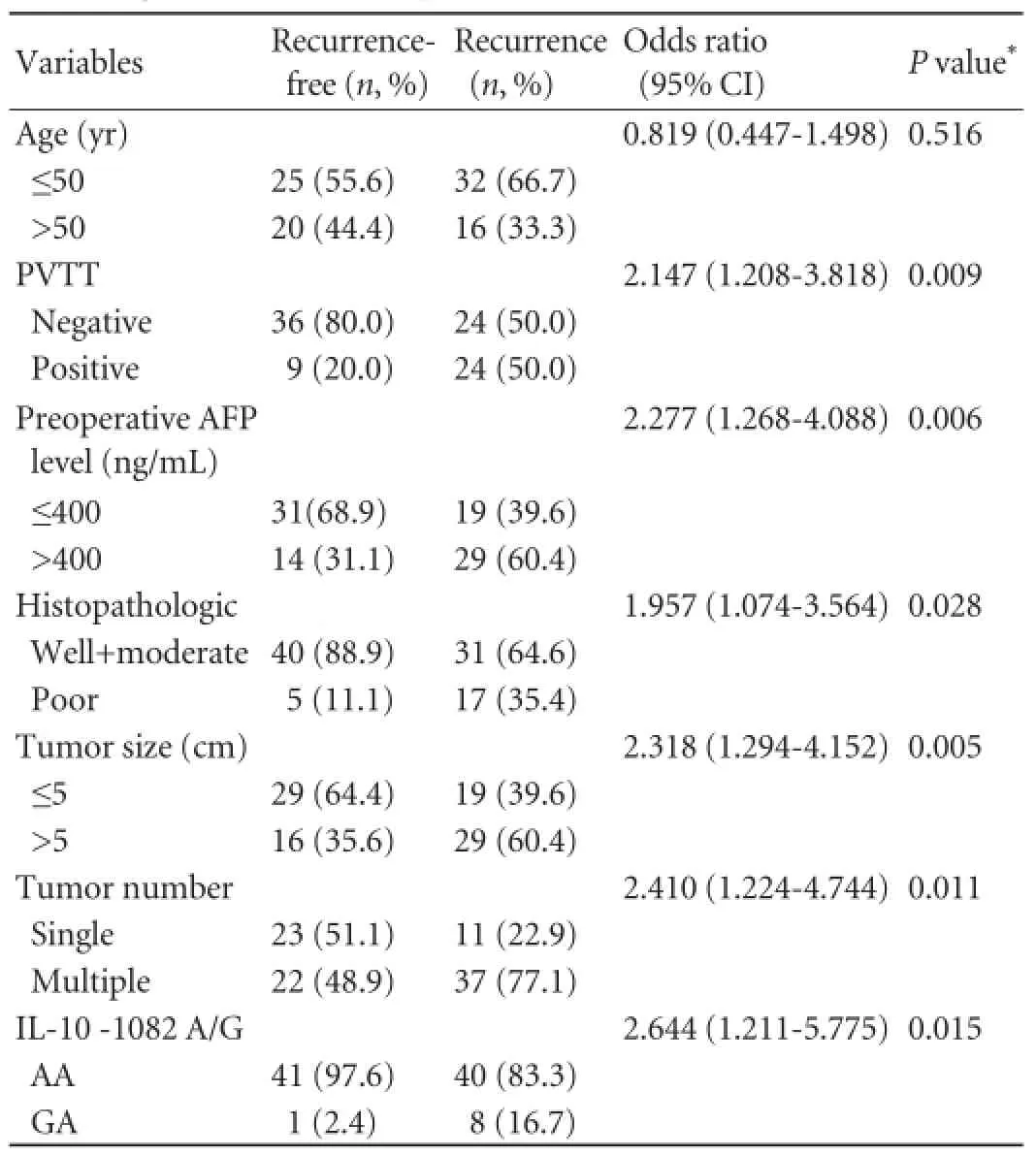
Table 4.Univariate analysis of main clinicopathological variables according to the absence or presence recurrent HCC after LT
The relationship between IL-10 -1082 genotype and clinicopathological parameters were also analyzed for excluding a plausible causative relationship. No signif i cance was found between AA genotype and GA genotype in the loci of IL-10 -1082 in distribution of clinicopathological parameters (allP>0.05, Table 3), indicating that these potential variables had no effect on the risk of tumor recurrence to IL-10 -1082 A/G genotype.
In addition, Kaplan-Meier survival curves according to IL-10 -1082 A/G genotypes showed a signif i cant association of RFS with different genotypes. Overall, the recurrence of HCC was identif i ed in 8/9 (88.9%) homozygous GA patients, and 40/81 (49.4%) heterozygous AA patients (Table 2). Patients with homozygous AA had a longer mean recurrence-free survival than heterozygous GA patients (23.5 vs 5.7 months,P=0.001). In the present study, univariate analysis demonstrated that besides clinicopathological variables including PVTT, preoperative AFP level, histopathological grading, tumor size and tumor number, IL-10 -1082 GA genotype (P=0.015) could signif i cantly predict the tumor recurrence after LT (Table 4). In the Cox model, IL-10 -1082 GA genotype, and other known prognostic factors such as PVTT, AFP level, histopathological tumor grading, tumor size and tumor number were included. However, multivariate analysis failed to reveal that GA genotype of IL-10 -1082 G/A was an independent indicator of recurrence (P>0.05, data not shown).
Discussion
Recent studies have demonstrated that individual's SNP genotype will provide additional information in the assessment of risk of tumor recurrence and individualized treatment modalities in HCC patients treated with LT.[3-5,21]In this study, we examined whether genetic variations of cytokine genes were of predictive value for HCC recurrence in patients undergoing LT.Obviously, our results do not support the role of SNPs of cytokine genes in prognosis of HCC following LT. To our knowledge, this is the fi rst investigation on the effect of cytokine gene polymorphisms on tumor recurrence in HCC patients treated with LT.
IL-10 has shown up as a potent immunosuppressive cytokine by down-regulating the expression of Th1 cytokines and costimulatory molecules and this gene is highly polymorphic. Many studies[22-24]investigated the potential diagnostic and prognostic role of IL-10 in HCC. High IL-10 levels indicated a worse disease-free survival in patients with resectable HCC.[23]A metaanalysis including seven studies suggested that IL-10 -592 A/C polymorphism may be associated with HCC among Asians.[24]At least three SNPs (-1082 A/G, -819 T/C, -592 A/C) in the promoter region of the IL-10 gene have been found so far. IL-10 -1082 A/G were most studied polymorphic loci. The correlation of the A to G substitution at position -1082 of the IL-10 gene promoter with increased transcription of IL-10 has been demonstratedin vitro.[25]Based on the promising data on IL-10, we ascertained the prognostic effect of IL-10 polymorphisms on recurrence after LT. Interestingly, our results showed a positive relationship between IL-10 -1082 AA genotype and recurrence-free after LT (Table 2). Whereas recent studies[18,19]showed that the IL-10 -1082 AA genotype was associated with unfavourable prognosis. Meanwhile, multivariate analysis failed to reveal that IL-10 -1082 GA genotype was an independent indicator of recurrence. We have not yet found a clear explanation for this result, but this is partly due to the small sample size and different background of the disease. Further investigations are required for the use of the SNPs of cytokine genes, including validation and analysis of additional retrospective and prospective cohorts.
So far, studies on the relationship between genomic polymorphisms of various cytokines and prognosis of human cancers resulted in controversial conclusions. Consistent with the results of some studies,[26-28]the present one did not fi nd positive results in the Chinese cohort. One reasonable explanation may be the complicated cytokine network, in which the cytokines have an effect on the expression of other factors in nonlinear relationships. Given the complexity of the cytokine network, the limited number of SNPs studied does not rule out the notion that other cytokine polymorphisms could act as candidate biomarkers of disease prognosis. On the other hand, this study has some limitations. First, the number of participants is relatively small, and thus failure to detect associations between some genotypes and HCC recurrence might be due to limited statistical power. Second, the study population almost unavoidably comprised patients with hepatitis B-induced HCC in the Han Chinese population, resulting in the different allele frequency due to ethnic background. Third, the number of samples detected for each polymorphism was not exactly equal because of a genotyping error or impaired DNA quality. Forth, the value of such study is decreased if it is limited only to the analysis of SNP association with a def i ned clinical observation and does not take into account the polymorphism gene effect on gene or protein expression. It is worth validating the result of the present study in the current patient cohort because of the improvement of criteria at our center for HCC patient selection for LT (Hangzhou criteria).[29]From the universal controversial studies, we propose a study with a larger cohort of patients, and well controlled analysis remains to be investigated. Further, research into the polymorphism gene effect on gene or protein expression is recommended in the future.
Because the prediction of gene expression-based tumor recurrence is largely based on direct assessment of the level in tumor tissue, SNP assay would be advantageous over that method. For example, SNP analysis is based on constitutional DNA and it can be made on peripheral blood samples instead of tumor tissues, which is theoretically applicable to every cancer patient and requires less invasive procedures. Moreover, there are few restrictions on sample collection and management because of the stability of DNA. Additionally, genotyping is less expensive and technically simpler than expression array analysis. Our studies including the present one illustrated that the systemic SNP array of cancer-related genes and combined analysis of HCC patients should be warranted.
In conclusion, this study is the fi rst one to investigate the association between cytokine gene polymorphisms and HCC recurrence after LT in a Chinese population. IL-10 -1082 polymorphisms might contribute to tumor recurrence in HCC patients although multivariate analysis failed to reveal the independently predictive power of IL-10 -1082 for tumor recurrence after LT.
Contributors:WLM and ZL proposed the study and wrote the fi rst draft. WLM and ZL contributed equally to the article. WLM, XJ and WBJ analyzed the data. All authors contributed to the design and interpretation of the study and to further drafts. ZSS is the guarantor.
Funding:This study was supported by grants from the National Natural Science Foundation of China (81101840), the National Natural Science Foundation of Innovation Group of China (81121002), and the National S&T Major Project of China (2012ZX10002017).
Ethical approval:This study protocol was approved by the Ethical Review Committee of the First Aff i liated Hospital, Zhejiang University School of Medicine.
Competing interest:No benef i ts in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
组织学生结合教材进行回答,并尝试着去理解文本的中心思想,使学生在阅读以及带着问题解读中更加深刻地理解作者对祖国文化的自豪感以及对劳动人民的热爱之情,进而帮助学生更好地理解。之后,教师可以鼓励学生自主借助网络来了解更多的石拱桥,并通过分析不同石拱桥的特点来形成那份自豪感,进而真正确保本节课的教学价值得以实现。
1 Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011;61:69-90.
2 Lau WY, Lai EC. Hepatocellular carcinoma: current management and recent advances. Hepatobiliary Pancreat Dis Int 2008;7:237-257.
3 Wu LM, Zhang F, Xie HY, Xu X, Chen QX, Yin SY, et al. MMP2 promoter polymorphism (C-1306T) and risk of recurrence in patients with hepatocellular carcinoma after transplantation. Clin Genet 2008;73:273-278.
4 Li XD, Wu LM, Xie HY, Xu X, Zhou L, Liang TB, et al. No association exists between E-cadherin gene polymorphism and tumor recurrence in patients with hepatocellular carcinoma after transplantation. Hepatobiliary Pancreat Dis Int 2007;6:254-258.
5 Li H, Xie HY, Zhou L, Wang WL, Liang TB, Zhang M, et al. Polymorphisms of CCL3L1/CCR5 genes and recurrence of hepatitis B in liver transplant recipients. Hepatobiliary Pancreat Dis Int 2011;10:593-598.
6 Howell WM, Rose-Zerilli MJ. Cytokine gene polymorphisms, cancer susceptibility, and prognosis. J Nutr 2007;137:194S-199S.
7 Alison MR, Nicholson LJ, Lin WR. Chronic inf l ammation and hepatocellular carcinoma. Recent Results Cancer Res 2011;185:135-148.
8 Ioana Braicu E, Mustea A, Toliat MR, Pirvulescu C, Könsgen D, Sun P, et al. Polymorphism of IL-1alpha, IL-1beta and IL-10 in patients with advanced ovarian cancer: results of a prospective study with 147 patients. Gynecol Oncol 2007;104:680-685.
10 Heneghan MA, Johnson PJ, Clare M, Ho S, Harrison PM, Donaldson PT. Frequency and nature of cytokine gene polymorphisms in hepatocellular carcinoma in Hong Kong Chinese. Int J Gastrointest Cancer 2003;34:19-26.
11 Wang Y, Kato N, Hoshida Y, Otsuka M, Taniguchi H, Moriyama M, et al. UDP-glucuronosyltransferase 1A7 genetic polymorphisms are associated with hepatocellular carcinoma in japanese patients with hepatitis C virus infection. Clin Cancer Res 2004;10:2441-2446.
12 Migita K, Miyazoe S, Maeda Y, Daikoku M, Abiru S, Ueki T, et al. Cytokine gene polymorphisms in Japanese patients with hepatitis B virus infection--association between TGF-beta1 polymorphisms and hepatocellular carcinoma. J Hepatol 2005;42:505-510.
13 Okamoto K, Ishida C, Ikebuchi Y, Mandai M, Mimura K, Murawaki Y, Yuasa I. The genotypes of IL-1 beta and MMP-3 are associated with the prognosis of HCV-related hepatocellular carcinoma. Intern Med 2010;49:887-895.
14 Snoussi K, Mahfoudh W, Bouaouina N, Ahmed SB, Helal AN, Chouchane L. Genetic variation in IL-8 associated with increased risk and poor prognosis of breast carcinoma. Hum Immunol 2006;67:13-21.
15 Kamali-Sarvestani E, Aliparasti MR, Atef i S. Association of interleukin-8 (IL-8 or CXCL8) -251T/A and CXCR2 +1208C/ T gene polymorphisms with breast cancer. Neoplasma 2007; 54:484-489.
16 Jeng JE, Tsai JF, Chuang LY, Ho MS, Lin ZY, Hsieh MY, et al. Tumor necrosis factor-alpha 308.2 polymorphism is associated with advanced hepatic fi brosis and higher risk for hepatocellular carcinoma. Neoplasia 2007;9:987-992.
17 Yu J, Zeng Z, Wang S, Tian L, Wu J, Xue L, et al. IL-1B-511 polymorphism is associated with increased risk of certain subtypes of gastric cancer in Chinese: a case-control study. Am J Gastroenterol 2010;105:557-564.
18 Bogunia-Kubik K, Mazur G, Wróbel T, Kuliczkowski K, Lange A. Interleukin-10 gene polymorphisms inf l uence the clinical course of non-Hodgkin's lymphoma. Tissue Antigens 2008;71:146-150.
19 Domingo-Domènech E, Benavente Y, González-Barca E, Montalban C, Gumà J, Bosch R, et al. Impact of interleukin-10 polymorphisms (-1082 and -3575) on the survival of patients with lymphoid neoplasms. Haematologica 2007;92:1475-1481.
20 Xie HY, Wang WL, Yao MY, Yu SF, Feng XN, Jin J, et al. Polymorphisms in cytokine genes and their association with acute rejection and recurrence of hepatitis B in Chinese liver transplant recipients. Arch Med Res 2008;39:420-428.
21 Wu LM, Xie HY, Zhou L, Yang Z, Zhang F, Zheng SS. A single nucleotide polymorphism in the vascular endothelial growth factor gene is associated with recurrence of hepatocellular carcinoma after transplantation. Arch Med Res 2009;40:565- 570.
22 Hsia CY, Huo TI, Chiang SY, Lu MF, Sun CL, Wu JC, et al. Evaluation of interleukin-6, interleukin-10 and human hepatocyte growth factor as tumor markers for hepatocellular carcinoma. Eur J Surg Oncol 2007;33:208-212.
23 Chau GY, Wu CW, Lui WY, Chang TJ, Kao HL, Wu LH, et al. Serum interleukin-10 but not interleukin-6 is related to clinical outcome in patients with resectable hepatocellular carcinoma. Ann Surg 2000;231:552-558.
24 Wei YG, Liu F, Li B, Chen X, Ma Y, Yan LN, et al. Interleukin-10 gene polymorphisms and hepatocellular carcinoma susceptibility: a meta-analysis. World J Gastroenterol 2011;17:3941-3947.
25 Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet 1997;24:1-8.
26 Talseth BA, Meldrum C, Suchy J, Kurzawski G, Lubinski J, Scott RJ. Lack of association between genetic polymorphisms in cytokine genes and disease expression in patients with hereditary non-polyposis colorectal cancer. Scand J Gastroenterol 2007;42:628-632.
27 Nahon P, Sutton A, Rufat P, Faisant C, Simon C, Barget N, et al. Lack of association of some chemokine system polymorphisms with the risks of death and hepatocellular carcinoma occurrence in patients with alcoholic cirrhosis: a prospective study. Eur J Gastroenterol Hepatol 2007;19:425-431.
28 Ognjanovic S, Yuan JM, Chaptman AK, Fan Y, Yu MC. Genetic polymorphisms in the cytokine genes and risk of hepatocellular carcinoma in low-risk non-Asians of USA. Carcinogenesis 2009;30:758-762.
29 Zheng SS, Xu X, Wu J, Chen J, Wang WL, Zhang M, et al. Liver transplantation for hepatocellular carcinoma: Hangzhou experiences. Transplantation 2008;85:1726-1732.
Received December 25, 2011
Accepted after revision May 31, 2012
AuthorAff i liations:Key Laboratory of Combined Multi-organ Transplantation, Ministry of Public Health; Key Laboratory of Organ Transplantation, Zhejiang Province; and Division of Hepatobiliary and Pancreatic Surgery, First Aff i liated Hospital, Zhejiang University School of Medicine, Hangzhou 310003, China (Wu LM, Zhou L, Xu J, Wei BJ, Chen J, Xu X, Xi B, Xie HY and Zheng SS)
Shu-Sen Zheng, MD, PhD, FACS, Key Laboratory of Combined Multi-organ Transplantation, Ministry of Public Health, First Aff i liated Hospital, Zhejiang University School of Medicine, Hangzhou 310003, China (Tel: 86-571-87236570; Fax: 86-571-87236466; Email: shusenzheng@zju.edu.cn)
© 2013, Hepatobiliary Pancreat Dis Int. All rights reserved.
10.1016/S1499-3872(13)60006-5
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Melanoma in the ampulla of Vater
- Hepatobiliary & Pancreatic Diseases International (HBPD INT)
- Salvage living-donor liver transplantation to previously hepatectomized hepatocellular carcinoma patients: is it a reasonable strategy?
- Clinical operational tolerance in liver transplantation: state-of-the-art perspective and future prospects
- Radiologic-histological correlation of hepatocellular carcinoma treated via pre-liver transplant locoregional therapies
- Outcomes of side-to-side conversion hepaticojejunostomy for biliary anastomotic stricture after right-liver living donor liver transplantation
