胰腺导管腺癌microRNA表达谱的研究
2012-11-07张晶赵晨燕刘清华余党会陈颖史敏倪灿荣朱明华
张晶 赵晨燕 刘清华 余党会 陈颖 史敏 倪灿荣 朱明华
·论著·
胰腺导管腺癌microRNA表达谱的研究
张晶 赵晨燕 刘清华 余党会 陈颖 史敏 倪灿荣 朱明华
目的应用microRNA(miRNA)高通量生物芯片筛选胰腺导管腺癌及癌旁组织差异表达的miRNA,分析其相关的靶基因。方法收集9例新鲜的胰腺导管腺癌和3例癌旁组织,运用标记713个miRNA的Agilent miRNA生物芯片筛选胰腺导管腺癌差异表达的miRNA,应用荧光实时定量PCR方法验证表达上调的miRNA。采用TargetScan 5.1和miRandaV5分析软件分析差异表达miRNAs的靶基因。结果miRNA芯片筛选出11个胰腺导管腺癌相关的差异表达的miRNA,其中miR-194*、miR-192*、miR-602、miR-194表达上调,miR-139-3p、miR-513a-5p、miR-630、miR-30c-1*、miR-887、miR-508-5p、miR-516a-5p表达下调。miR-192、miR-194及其同源体的表达在31例胰腺癌组织中得到验证。经软件分析,miR-192靶基因有ZEB2、CXCL-2、EEF1A1、ERCC3,miR-192*靶基因有DCC、SMAD4、FAS,miR-194靶基因有DACH1、IGSF11、PTPN2、RBBP4,miR-194*靶基因有CD40LG、CIDEB、FHL1。结论胰腺导管腺癌存在11个表达差异的miRNA,这些miRNA可能与胰腺导管腺癌的发生、发展有关。
胰腺肿瘤; 微RNAs; 微阵列分析; 基因表达谱
MicroRNA(miRNA)是近年来发现的一类长度为19~23 nt的、广泛存在于动植物体内的、非编码单链RNA。miRNA通过作用于其靶mRNA,降解靶基因或抑制其翻译,发挥负调控作用。研究表明[1-6],miRNA参与生命过程中一系列的重要进程,包括早期发育、细胞增殖及分化、凋亡和免疫调节等,还具有癌基因和抑癌基因的作用。在不同类型的肿瘤中有其特异的miRNA表达谱。本研究采用miRNA芯片技术筛选胰腺导管腺癌与癌旁正常胰腺组织差异表达的miRNA,并分析其相关靶基因。
材料与方法
一、组织标本
收集2007年至2008年长海医院外科切除的9例胰腺癌组织标本及3例配对的癌旁正常胰腺标本送上海生物芯片有限公司行miRNA芯片检测,其中男性4例,女性5例,年龄47~75岁。另取2009年3月至2010年5月手术切除的31例胰腺癌及配对的癌旁新鲜组织标本,液氮冷冻后置-80℃保存,行实时定量PCR;其中男性15例,女性16例,年龄45~77岁。所有患者术前均未进行抗肿瘤治疗,均签署知情同意书,并经第二军医大学长海医院伦理委员会批准。所有标本经病理检查均证实为胰腺导管腺癌。
二、miRNA芯片检测
应用miRNA抽提试剂盒(Ambion, AM1560)抽提新鲜组织的mRNA,并富集miRNA。应用miRNA标记和杂交试剂盒(Agilent 5190-0408)对miRNA进行CY3荧光标记。miRNA微阵列基因芯片和质控探针由上海生物芯片有限公司提供(Agilnent公司产品),其miRNA探针序列信息来自于SangermiRBase Release10.0版本数据库(http://microrna. sanger. ac. uk/sequences/),涵盖了Sanger数据库中713个miRNA探针。每张芯片都设计了与实验样品无同源性的核酸序列作为质控RNA。组织标本预处理及探针杂交按Agilent芯片实验操作步骤进行。设空白对照及采用与实验样品无同源性的核酸序列探针的阴性对照组。芯片的每个探针杂交位点至少有3个重复。
采用Agilent扫描仪扫描采集杂交图像,采用Feature Extraction软件读取图像数据,最后采用Feature Extraction进行处理分析,得到的原始数据通过VSN(variance stabilizing normalization)方法进行归一化。
三、荧光实时定量PCR
应用miRNA抽提试剂盒抽提31例配对的胰腺导管腺癌和癌旁胰腺组织miRNAs,应用TaqMan microRNA reverse transcription试剂盒将miRNAs逆转录成cDNA,应用TaqMan microRNA assays检测miRNAs的表达。PCR反应条件:95℃ 30 s,95℃ 5 s,60℃ 34 s,40个循环。以U6 SnRNA作为内参照,采用△CT方法计算miRNA的相对表达量。
四、差异miRNA靶基因及其相关疾病分析
采用生物信息学分析方法。应用TargetScan5.1和miRandaV5分析软件预测并初步分析差异表达miRNAs的靶基因,并分析与其相关的疾病。
五、统计学分析
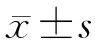
结 果
一、胰腺导管腺癌组织中差异表达的miRNA
miRNA芯片杂交信号强度由绿-黑-红色依次增加。胰腺导管腺癌组织中差异表达≥1.2倍的miRNA共12个,其中5个上调,为miR-194*、miR-192*、miR-602、miR-801、miR-194;7个下调,为miR-139-3p、miR-513a-5p、miR-630、miR-30c-1*、miR-887、miR-508-5p、miR-516a-5p(表1)。因miR-801被证实不是真正的miRNA,而是一个U11RNA的片段,故在聚类分析中被剔除(图1)。上调最高的miRNA为miR-194*,上调倍数为2.38倍。
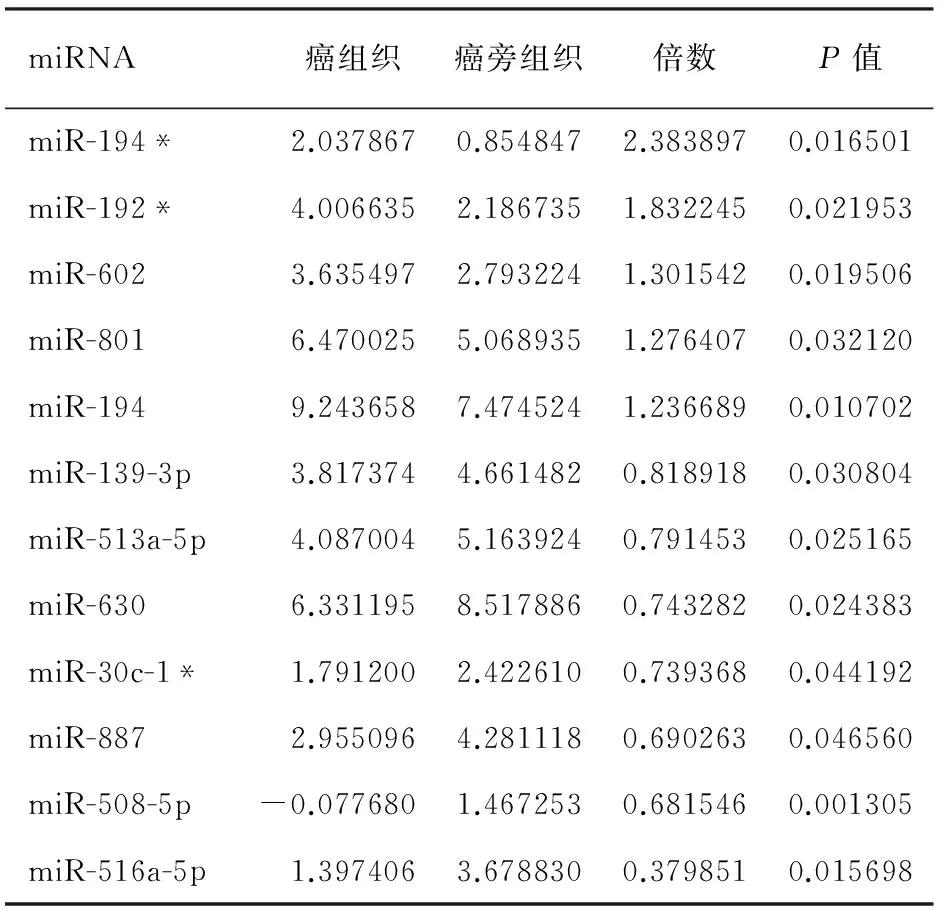
表1 胰腺导管腺癌中差异表达的miRNAs
注:带*号的miRNA与其相应的miRNA来自同一前体

图1 差异表达的miRNAs聚类分类图
二、胰腺癌组织miR-192、miR-192*、miR-194和miR-194*表达的验证
31例胰腺导管腺癌和配对癌旁胰腺组织的miR-192、miR-192*、miR-194和miR-194*表达均显著上调(P值均<0.05,表2)。
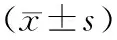

miRNA胰腺癌组织癌旁组织P值miR-192-0.66±4.08-5.49±7.140.001miR-192*-6.63±4.97-8.98±3.510.010miR-194-1.56±4.15-6.51±7.790.001miR-194*-9.35±5.51-11.95±2.860.022
三、差异表达miRNA的靶基因及其相关疾病
miR-192靶基因有ZEB2、CXCL-2、EEF1A1,miR-192*靶基因有DCC、SMAD4、FAS,miR-194靶基因有DACH1、IGSF11、PTPN2、RBBP4,miR-194*靶基因有CD40LG、CIDEB、FHL1。相关的疾病见表3。
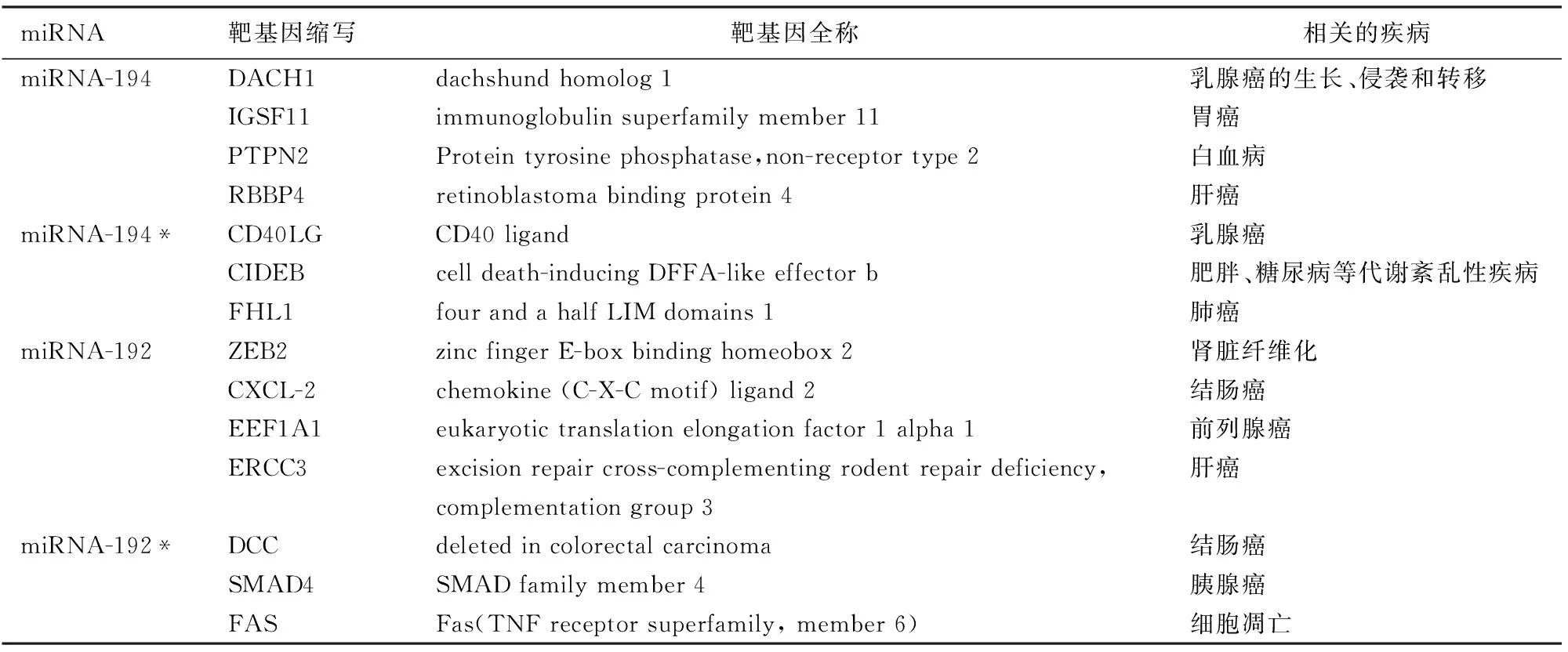
表3 差异表达miRNA的靶基因及相关疾病
讨 论
人类约50%已知的miRNAs基因定位于与肿瘤相关的染色体区,提示其在肿瘤发生中具有重要作用[6]。在慢性淋巴细胞性白血病、儿童Burkitt淋巴瘤、弥漫性大B细胞淋巴瘤、肺癌和乳腺癌等恶性肿瘤中,miRNAs表达均发生显著变化[7-14],并可抑制或促进肿瘤的发生和发展。2006年Volinia等[15]报道,胰腺癌中异常高表达的miRNA有55个,低表达的miRNA有2个。随后,Lee、Bloomston及Szafanska等[16-18]检测了胰腺癌、癌旁组织、慢性胰腺炎组织中miRNA表达谱,发现胰腺癌中异常高表达的miRNA有miR-155、miR-21、miR-221、miR-222、miR-196a等,异常低表达的有miR-181、miR-148、miR-216和miR-217等。Szafanska等[18]还发现miRNA表达谱可用于鉴别胰腺癌、慢性胰腺炎和正常胰腺。
本研究应用Agilent miRNA芯片筛选出胰腺导管腺癌表达上调的miRNA 4个,下调的miRNA 7个,其中miR-192*和miR-194*上调大于1.8倍。通过胰腺癌组织标本的检测,证实了miRNA芯片的结果。
Mees等[19]在miRNA上游转录因子的研究中发现,高侵袭、高转移的胰腺癌细胞株中miR-194的水平明显高于侵袭力和浸润、转移能力较差的胰腺癌细胞株。Meng等[20]发现miR-194在肝上皮细胞中高表达,它可作为肝上皮细胞的标志物,并能抑制小鼠体内肝癌细胞的转移。miR-194也表达于胃肠道和肾脏,但肝脏枯否细胞和星状细胞无高表达。
通过靶基因预测软件检索,miR-194的靶基因有DACH1。DACH1蛋白是黑腹果蝇dachshund(Dac)基因在脊椎动物中表达的类似物,是一种新型的抑癌基因,它通过阻断肿瘤上皮细胞的DNA合成、抑制肿瘤基质层中肿瘤集落的形成和肿瘤细胞的有丝分裂达到阻止肿瘤细胞增殖的目的。有研究[21]证明SIP1为miR-192的靶基因。Kato等[22-23]研究发现,miR-192在纤维化的肾脏高度特异性表达,促进细胞外基质(ECM)蛋白的蓄积;并通过双荧光素检测确定SIPl的3′UTR为miR-192调控靶点,证明miR-192通过mRNA降解途径降低SIPl蛋白的表达。我们的结果并没有显示SIP1为miR-192的靶基因,这可能与生物信息学分析的方法不同、生物信息学分析与实验检测存在一定的差异有关。
[1] Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans.Nature, 2000, 403:901-906.
[2] Brennecke J, Hipfner DR, Stark A, et al. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila.Cell, 2003, 113:25-36.
[3] Xu P, Vernooy SY, Guo M, et al. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol, 2003, 13:790-795.
[4] Dostie J,Mourelatos Z,Yang M,et al.Numerous microRNPs in neuronal cells containing novel microRNAs.RNA,2003,9:180-186.
[5] Chen CZ, Li L, Lodish HF, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science, 2004,303:83-86.
[6] Calin GA, Sevignani C,Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA, 2004, 101:2999-3004.
[7] Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13ql4 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA,2002,99:15524-15529.
[8] He L, Thomson JM ,Hemann MT, et al. A microRNA palycistron as a potential human oncogene.Nature, 2005,435:828-833.
[9] Hanck L, von Harsdorf R. E2F transcription factors and pRb pocket proteins in cell cycle progression. Methods Mol Biol, 2005, 296:239-245.
[10] van den BergA, Kroesen BJ, Kooistra K, et al. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer, 2003, 37:20-28.
[11] Metzler M, Wilda M, Busch K, et al. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma.Genes Chromosomes Cancer, 2004, 39:167-169.
[12] Eis PS, Tam W, Sun L, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA, 2005, 102:3627-3632.
[13] Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res, 2004, 64:3753-3756.
[14] Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res, 2005, 65:9628-9632.
[15] Volinia S, Calin GA, Liu CG,et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA, 2006, 103:2257-2261.
[16] Lee EJ, Gusev Y, Jiang J,et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer, 2007, 120:1046-1054.
[17] Bloomston M, Frankel WL, Petrocca F,et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA, 2007, 297:1901-1908.
[18] Szafranska AE, Davison TS, John J,et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene, 2007, 26:4442-4452.
[19] Mees ST, Mardin WA, Wendel C,et al. EP300-a miRNA-regulated metastasis suppressor gene in ductal adenocarcinomas of the pancreas. Int J Cancer, 2010, 126:114-124.
[20] Meng Z, Fu X, Chen X,et al. miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology, 2010, 52:2148-2157.
[21] Wang B, Herman-Edelstein M, Koh P, et al. E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-beta. Diabetes, 2010, 59:1794-1802.
[22] Kato M, Zhang J, Wang M,et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA, 2007, 104:3432-3437.
[23] Kato M, Arce L, Wang M,et al. A microRNA circuit mediates transforming growth factor-β1 autoregulation in renal glomerular mesangial cells. Kidney Int,2011,80:358-368.
MicroRNAprofilesinpancreaticductaladenocarcinoma
ZHANGJing,ZHAOChen-yan,LIUQing-hua,YUDang-hui,CHENYing,SHIMin,NICan-rong,ZHUMing-hua.
DepartmentofPathology,ChanghaiHospital,SecondMilitaryMedicalUniversity,Shanghai200433,China
ZHUMing-hua,Email:mhzhu2000@hotmail.com
ObjectiveTo study the differentially expressed microRNA (miRNA) between pancreatic ductal adenocarcinoma (PDAC) and para-cancerous tissues, and determine related target genes.MethodsNine fresh PDAC tumor tissues and 3 adjacent normal pancreatic tissues were collected, then Agilent miRNA microarray with 713 miRNA loci was used to identify the differentially expressed miRNA. Real-time PCR method was applied to verify the up-regulated miRNA. TargetScan 5.1 and miRandaV5 software were used to analyze the related target genes.ResultsmiRNA microarray identified 11 PDAC related miRNAs, among them, the expressions of miR-194*, miR-192*, miR- 602, miR-194 were up-regulated, while the expressions of miR-139-3p, miR-513a-5p, miR-630, miR-30c-1*, miR-887, miR-508-5p, miR-516a-5p were down-regulated. The expressions of miR-192, miR-194 and their homolog were verified in 31 PDAC tumor tissues. After software analysis, it was found that target genes of miR-192 were ZEB2, CXCL-2, EEF1A1, ERCC3, and target genes of miR-192* were DCC, SMAD4, FAS, and target genes of miR-194 included DACH1, IGSF11, PTPN2, RBBP4, while target genes of miR-194* included CD40LG, CIDEB, FHL1.ConclusionsEleven differentially expressed miRNAs are present in PDC, and they may be involved in the occurrence and development of PDC.
Pancreatic neoplasms; MicroRNA; Microarray analysis; Gene expression profiling
10.3760/cma.j.issn.1674-1935.2012.05.007
国家自然科学基金(30770996;81172310)
200433 上海,第二军医大学长海医院病理科(张晶、刘清华、余党会、陈颖、史敏、倪灿荣、朱明华);复旦大学附属妇产科医院病理科(赵晨燕)
共同第一作者:赵晨燕
朱明华,Email:mhzhu2000@hotmail.com
2012-06-13)
(本文编辑:屠振兴)
