Genetic differentiations between randomly and selectively bred pig populations in Yunnan, China
2011-12-25QUKaiXingWUGuiShengGOUXiaoYANDaWeiLIANLinShengMumtazBaigZHANGYaPing
QU Kai-Xing, WU Gui-Sheng, GOU Xiao, YAN Da-Wei, LIAN Lin-Sheng, Mumtaz Baig, ZHANG Ya-Ping
(1. Yunnan Key Laboratory of Molecular Biology of Domestic Animal & Laboratory of Molecular Evolution and Genome Diversity, Kunming Institute of Zoology, the Chinese Academy of Sciences, Kunming 650223, China; 2. Laboratory for Conservation and Utilization of Bio-resource, Yunnan University, Kunming 650091, China; 3. Yunnan Academy of Grassland and Animal Science, Xiaoshao, Kunming 650212, China; 4. Kunming Institute of Botany, the Chinese Academy of Sciences, Kunming 650204, China; 5. College of Animal Science and Technology, Yunnan Agricultural University, Kunming 650201, China; 6. Department of Zoology, Govt Vidarbha Institute of Science and Humanities, Amravati 444604, India)
Genetic differentiations between randomly and selectively bred pig populations in Yunnan, China
QU Kai-Xing1,2,3, WU Gui-Sheng1,4, GOU Xiao5, YAN Da-Wei5, LIAN Lin-Sheng5, Mumtaz Baig6, ZHANG Ya-Ping1,2,*
(1.Yunnan Key Laboratory of Molecular Biology of Domestic Animal & Laboratory of Molecular Evolution and Genome Diversity,Kunming Institute of Zoology,the Chinese Academy of Sciences,Kunming650223,China; 2.Laboratory for Conservation and Utilization of Bio-resource,Yunnan University,Kunming650091,China; 3.Yunnan Academy of Grassland and Animal Science,Xiaoshao,Kunming650212,China; 4.Kunming Institute of Botany,the Chinese Academy of Sciences,Kunming650204,China; 5.College of Animal Science and Technology,Yunnan Agricultural University,Kunming650201,China; 6.Department of Zoology,Govt Vidarbha Institute of Science and Humanities,Amravati444604,India)
To assess the genetic diversity between randomly and selectively bred populations, we sequenced 438 bp of the mitochondrial DNA control region from 102 pigs. These samples represented four native pig breeds, one nucleus and one conservation herd from Yunnan, China. Twenty haplotypes with sixteen polymorphic sites were identified. The number of haplotypes in the nucleus herd of Saba pig and the conservation herd of Banna miniature pig were restricted to three and one, respectively, while the randomly bred pig populations exhibited over six haplotypes. Notably, haplotype diversity in randomly bred populations was significantly greater than the selectively bred populations (h=0.732 vs. 0.425 and 0, exact test,P≤0.0036). These findings demonstrate that selective breeding generated low genetic diversity compared to randomly bred pig breeds. A timely intervention and well programmed breeding approach would stop further genetic diversity reduction in the nucleus and conservation herds of native pig breeds. Otherwise, selective breeding would dramatically reduce genetic diversity in only several years, indicating that sharp contradictions exist between breeding, conservation and genetic diversity. Genetic relationships are discussed based on net genetic distances among pig populations.
Yunnan pig breeds; Genetic diversity; Randomly bred population; Selectively bred population
1 Material and Methods
1.1 Breeding background of native Yunnan pig breeds
A breeding herd of Saba pigs was established in 1991 at the Pig Breeding Farm of Chuxiong Prefecture, consisting of four boars and twenty sows from Luquan County in Yunnan. In 1993, fifteen boars and fifty-five sows from Shuangbai County in Yunnan were added to the breeding herd. After observation and testing, eight boars and thirty-five sows were selected to remain in the breeding herd. In 1996, the nucleus herd was established, in which one boar and four sows from Yao’an County were introduced. Systematic selective breeding was used and high quality animals were selected for repeated reproduction. The nucleus herd was opened periodically. Random mating and organized mating were equally practiced. A sow yielded for every generation annually (Lian et al, 2003). Similarly, in the conservation herd of Banna miniature pigs from the Pig Breeding Farm of Xishuangbanna Prefecture, eight boars and forty sows were introduced from the remote countryside in 1987. A proportion of the population was moved to Yunnan Agricultural University of Kunming in 1990 as part of a conservation program. Systematic breeding was also conducted on the Banna miniature pig. The herd was closed after the base population was grouped, and randomly mating was practiced, with the exception of sib mating. Closed herd breeding was subsequently carried out (Lian et al, 2003). The initial base population of the Baoshan pigs consisted of three boars and forty-seven sows brought from mountainous villages of Shidian County in Baoshan Prefecture. In 2001, six boars and thirty-seven sows were introduced from Longyang District, Shidian County, and Changning County to join the base population. The nucleus herd was opened periodically (Lian, 2005) and an open nucleus breeding system was also applied to the Baoshan pigs. High quality animals were retained and used to reproduce repeatedly.
Randomly bred populations were comprised of native pig breeds from the remote countryside. These included Saba pigs from Lufeng County, Diqing Tibetan pigs from Zhongdian County, and Banna miniature pigs from Xishuangbanna Prefecture (Fig. 1).
1.2 DNA isolation, PCR amplification and automated DNA sequencing
Blood or muscle tissue samples were collected from pigs from different regions of Yunnan Province (Fig. 1). We randomly sampled inter-families, and focused on morphological characteristics to exclude hybrids. The DNA was isolated and purified (Huang et al, 1999), and 20-50 ng genomic DNA was used as a template for polymerase chain reaction (PCR) amplification of the mtDNA control region. The primers mitL99 (5'-15336CCCAAAGCTGAAATTCTAAA-3') and mitH451 (5'-15822GGTGAGATGGCCCTGAAGTAAG-3') were designed (Takeda et al, 1995) using a reference sequence ofSus scrofa(Accession nos. AF304201).
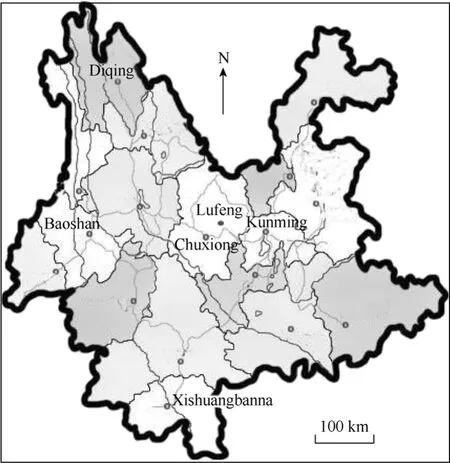
Fig. 1 Map of Yunnan, China, showing sampled prefectures/counties
The PCR amplifications were performed in 50 µL of 2.5 mmol/L of MgCl2, 200 µmol/L of each dNTPs, 1 µmol/L of each primer, and 0.25 U ofTaqpolymerase. Amplification was carried out using 35 cycles of denaturation for 3 min at 95 °C and extended by 1 min at 94 °C. Annealing was done for 1 min at 60 °C, followed by 1 min at 72 °C, and a final extension of 5 minutes at 72 °C. The PCR products were purified using spin columns (Watson BioTechnologies Inc., Shanghai). Purified amplicons were sequenced directly for both the DNA strands using the ABI PRISM BigDyeTMTerminator Cycler Sequencing Ready Reaction Kit (Applied Biosystems, CA USA). The resulting raw sequences were checked using DNAStar 5.0 (DNAStar Inc., Madison, WI) and submitted to GenBank (Accession numbers: AY178254~AY178265, AY178267~AY178274). Polymorphic sites and haplotypes were determined using MEGA software, Version 2.1 (Kumar et al, 2001) and DNASP software, Version 3.00 (Rozas & Rozas, 1999). Population diversity, population differentiation and exact test were performed using Arlequin Version 2.00 (Schneider et al, 2000). Nucleotide diversity and net genetic distance for inter- and intra-populations were computed using DNASP, Version 3.00 (Rozas & Rozas, 1999). A neighbor-joining tree was constructed by the net genetic distance matrix among populations using MEGA software, Version 2.1 (Kumar et al, 2001), under the Kimura 2-parameter substitution model.
2 Results
2.1 Genetic diversity of Yunnan pig breeds
Four hundred and thirty-eight base pairs of mtDNA control region were identified by comparison with the control region ofSus scrofa,GenBank (Accession nos. AF304201). Twenty haplotypes, including sixteen polymorphism sites (Tab. 1 and Tab. 2), were detected, of which five were singleton polymorphic sites and eleven were parsimony informative sites. Fifteen transition and two transversion substitutions were identified. No deletion/insertion was found in the entire 102 sequence dataset. Haplotype H23 was dominant (83.3%) in both the conservation herd of Banna miniature pig and the nucleus herd of Saba pig. The same maximum haplotype was shared (40.20%) in the samples. There were seven haplotypes (H3, H6, H7, H8, H16, H18, and H21) with single occurrence (0.98%). Some specific haplotypes, such as H7 in Baoshan pigs, and H8 and H13 in Diqing Tibetan pigs, were also found. Further, haplotypes H16, H17, H18, and H19 and H3 and H4 were found in the randomly bred Banna miniature pig population and the Saba pig population, respectively. Two more haplotypes (H21 and H22) were restricted exclusively to the nucleus herd of Saba pig (Tab. 2). The numbers of haplotypes in the conservation herd of Banna miniature pig and nucleus herd of Saba pig were one and three, respectively. The number of haplotypes found in the randomly bred populations ranged from six to ten. Haplotype diversity (h) and nucleotide diversity (π) in the randomly bred populations were significantly higher than the selectively bred populations (P<0.01) (Tab. 3).
2.2 Genetic relationship among six pig populations
Nucleotide diversity and net genetic distance between inter- and intra-populations are shown in Tab. 4. A neighbor-joining tree constructed by net genetic distances demonstrated the genetic relationship among the six pig populations. The nucleus herd of Saba pig and the conservation herd of Banna miniature pig initially clustered together; then they clustered with randomly bred population of Saba pig and Baoshan pig. All these pigs had similar morphological and physiological characteristics and belonged to Southwest-type of Chinese pig breeds (Lian et al, 2003). There was, however, very distinct genetic distance between the randomly bred Banna miniature pig population and the other populations (Fig. 2).
3 Discussion
The evolution rate of the mitochondrial DNA control region is several times higher than those of other mtDNA regions and nuclear genes. Because of this, the
D-loop region has been used extensively to study relationships among populations (Brown et al, 1979; Watanabe et al, 1986, 1999; Lin et al, 1999; Giuffra et al, 2000; Luikart et al, 2001). The sequences of 438 bp of the mtDNA control region in 102 pigs revealed twenty haplotypes with sixteen polymorphic sites. The number of haplotypes in Baoshan pig and Diqing Tibetan pig, and randomly bred Saba and Banna miniature pigs were 10, 9, 7, and 6, respectively (Tab. 2). Similarly, the number of haplotypes in the nucleus herd of Saba pig and the conservation herd of Banna miniature pig were confined to three and one (h=0.732 via 0.425 and zero), respectively. Low haplotype diversity indicated that long-term intensive breeding has significantly reduced diversity in the selectively bred pig populations (exact test,P≤0.0036). It was suggested that randomly bred populations possessed high diversity. But the population differentiation between the nucleus herd of Saba pig and the conservation herd of Banna miniature pig was not significant (P=0.688). Historically, pig breed resources in Yunnan Province were formed due to the wider fields, greatly different climates and people’s living customs (Lian et al, 2003). Over the years geographical, climatic, and cultural factors have restricted the gene flow between pig populations and resulted in the inheritance of few selected variable sites (or haplotypes) in subsequent generations. Compared to the nucleus herd of Saba pig, gene flow has been restricted in the base population of Baoshan pig. This is mainly due to the grouping of the Baoshan pig for breeding in 1998 and sampling in 1999, which led to the fixation of many extant haplotypes in the region. After seven years of
selection, some lineages were eliminated after only a few generations of breeding in the nucleus herd of Saba pig, which led to the loss of some haplotypes before breeding. Moreover, during the fifteen-year conservation program of Banna miniature pig, haplotype diversity has reduced to a single lineage (Lian et al, 2003). Haplotype diversity in the nucleus herd of Saba pig was 0.425, significantly lower than the lowest haplotype diversity (0.732) found in the randomly bred population (Tab. 3) (exact test, ofP<0.01). Furthermore the Diqing Tibetan pig exhibited a haplotype diversity of 0.925, double that found in the nucleus herd of Saba pig. Compared to the four randomly bred populations, nucleotide diversity was similar for the conservation and nucleus herds and fell in the range of 0.00205-0.00595 (Tab. 3). Our results clearly suggest that human induced factors have generated great differences in diversity among native pig breeds. Our study also reinforces the finding that population fragmentation and captive breeding greatly reduces diversity, as reported in blue chaffinch, Hector’s dolphin, cheetah, and sea otter (Menotti-Raymond & O’Brien, 1993; Pestano et al, 2000; Pichler & Baker, 2000; Freeman et al, 2001; Larson et al, 2002; Yildiz et al, 2002).

Tab. 1 The sixteen polymorphic sites of mitochondrial DNA control region

Tab. 2 Distribution of haplotypes in the pig populations

Tab. 3 The diversity indices in the six pig populations from Yunnan

Tab. 4 Estimates of interpopulational (dxy), intrapopulational (dx or dy), and net nucleotide diversity (dA) among the six pig populations in Yunnan
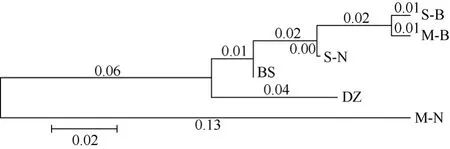
Fig. 2 N-J tree constructed by net genetic distances among the six populations
It is interesting to note that two bred populations (the nucleus herd of Saba pig and the conservation herd of Banna miniature pig) were initially clustered together in the N-J tree. We concluded the reasons were: (1) It was possible that mitochondrial haplotypes were linked with or related to some husbandry economic traits (Bell et al, 1985; Toelle et al, 1986; Brown et al, 1988; 1989), and the two pig populations may act to increase genetic homogeneity by paying more attention to litter size improvement for undertaking breeding programmes (Lian et al, 2003) and keeping captive populations (Nei, 1987; Menotti-Raymand & O’Brien, 1993; Freeman et al, 2001). Litter size improvement was mainly emphasized in the early period of selection and grouping (Lian et al, 2003) and dams possessing high litter size were selected and kept. At the same time, maternally inherited mitochondrion and special haplotypes were reasonably fixed in the populations and the frequency of special haplotypes increased. (2) Founder effect. The populations that first grouped and were selected accommodated a wider haplotype-pool, such as observed in the Baoshan pigs. Consequently, we traced the frequency fluctuation after some generations. Furthermore, dams possessing high litter size kept the same haplotypes as their mother and grandmother, and the haplotypes were inclined to simplification. Saba and Baoshan pigs, which belong to the Southwest type of Chinese pig breeds according to morphological and physiological characteristics, clustered together with the bred populations. We concluded that gene exchange with the Diqing Tibetan pig during their formation had occurred because of sharing four haplotypes (Data not shown). The Banna miniature pig was located furthest away in the southern area of Yunnan Province, and also showed the furthest relationship with other Yunnan native pig breeds. The neighbor-joining tree showed that the relationship between Yunnan native pig breeds was consistent with their geographical distribution and physiological characteristics.
In regards to domestic animal breeding programs, farmers have pursued genotype homozygosity to obtain expected heterosis. Meeting some productive requirements, however, comes at the cost of diversity. A possible link between mitochondrial haplotypes with some economic traits in animal husbandry couldn’t be excluded (Bell et al, 1985; Brown et al, 1988,1989; Mannen et al, 1998,2003). Improvement in litter size was emphasized in early selection and grouping, with higher litter size sows selected and kept (Lian et al, 2003). Mitochondrial haplotypes inherited maternally to the next generation undergo expansion in the offspring easily. So, timely intervention and well programmed breeding approaches can stop further reduction in the genetic diversity of nucleus and conservation herds of native pig breeds. Hence, animals that were first grouped and selected overlaid more haplotypes. The Baoshan pig exhibited a similar pattern, and hence it became necessary to estimate haplotype frequency after only a few generations of breeding. Taking into account the single genetic marker, probing by Y-chromosomal and autosomal markers in the future will clarify the genetic differentiation associated with native pig breeds of Yunnan.
Bell BR, McDaniel BT, Robison OW. 1985. Effects of cytoplasmic in heritance on production traits of dairy cattle [J].J Dairy Sci,68: 2038-2051.
Brown DR, DeNise SK, McDaniel RG. 1988. Mitochondrial respiratory metabolism and performance of cattle [J].J Anim Sci,66: 1347-1354.
Brown DR, Keohler CM, Lindberg GL, Freeman AE, Mayfield JE, Myers AM, Schutz MM, Beitz DC. 1989. Molecular analysis of cytoplasmic genetic variation in Holstein cows [J].J Anim Sci,67: 1926-1932.
Brown WM, George MJ, Wilson AC. 1979. Rapid evolution of animal mitochondrial DNA [J].Proc Natl Acad Sci USA,76: 1967-1971.
Brown WM, Prager EM, Wang A, Wilson AC. 1982. Mitochondrial DNA sequence of primates: Tempo and mode of evolution [J].J Mol Evol,18: 225-239.
Disotell TR, Honeycutt RL, Ruvolo M. 1992. Mitochondrial DNA phylogeny of the old-world monkey tribe Papionini [J].Mol Biol Evol,9: 1-13.
Freeman AR, MacHugh DE, McKeown S, Walzer C, McConnell DJ, Bradley DG. 2001. Sequence variation in mitochondrial DNA control region of African cheetahs (Acinonyx jubatus) [J].Heredity,86: 355-362.
Giuffra E, Kijas JMH, Amarger V, Carlborg O, Jeon JT, Andersson L. 2000. The origin of the domestic pig: Independent domestication and subsequent introgression [J].Genetics,154: 1785-1791.
Huang YF, Shi XW, Zhang YP. 1999. Mitochondial genetic variation in Chinese pigs and wild boars [J].Biochem Genet,37: 335-343.
Jone GF. 1998. Genetic aspects of domestication, common breeds and their origin [M]// Ruvinsky A, Rothschild MF. The Genetics of the Pig. Oxon, UK: CAB International, 17-50.
Kijas JMH, Anderson L. 2001. A phylogenetic study of the origin of the domestic pig estimated from the near-complete mtDNA genome [J].J Mol Evol,52: 302-308.
Kim KI, Lee JH, Li K, Zhang YP, Lee SS, Gongora J, Moran C. 2002. Phylogenetic relationships of Asian and European pig breeds determined by mitochondrial DNA D-loop sequence polymorphism [J].Anim Genet,33: 19-25.
Kumar S, Tamura K, Jakobsen IB, Nei M. 2001. MEGA2: Molecular evolutionary genetics analysis [J]. Bioinformatics, 17: 1244-1245.
Larson S, Jameson R, Etnier M, Fleming M, Bentzen P. 2002. Loss of genetic diversity in sea otters (Enhydra lutris) associated with the fur trade of the 18th and 19th centuries [J].Mol Ecol,11: 1899-1903.
Lian LS. 2005. Baoshan pig: Protection and utilization of Yunnan native pig breeds [J].Anim Sci Vet Med, 22(9): 72-73. [连林生. 2005.云南地方猪种保护与利用之一—— 保山猪. 动物科学与动物医学, 22(9): 72-73.]
Lian LS, Xu BM, Lu SX. 2003. Genetic Diversity and Sustainable Utilization of Yunnan Native Pig Breeds [M]. Kunming: Yunnan Science and Technological Press. [连林生,徐宝明,鲁绍雄. 2003.云南土著猪种遗传多样性及其可持续利用 [M]. 昆明: 云南科技出版社.]
Lin CS, Sun YL, Liu CY, Yang PC, Chang LC, Cheng IC, Mao SJ, Huang MC. 1999. Complete nucleotide sequence of pig (Sus scrofa) mitochondrial genome and dating evolutionary divergence within artiodactyls [J].Gene,236: 107-114.
Luikart G, Gielly L, Excoffier L, Vigne JD, Bouvet J, Taberlet P. 2001. Multiple maternal origins and weak phylogeographic structure in domestic goats [J].Proc Natl Acad Sci USA,98: 5927-5932.
Mannen H, Kojima T, Oyama K, Mukai F, Ishida T, Tsuji S. 1998. Effect of mitochondrial DNA variation on carcass traits of Japanese Black cattle [J].J Anim Sci,76: 36-41.
Mannen H, Morimoto ML, Oyamat K, Mukai F, Tsuji S. 2003. Identification of mitochondrial DNA substitutions related to meat quality of Japanese Black cattle [J].J Anim Sci,81: 68-73.
Menotti-Raymond M, O’Brien SJ. 1993. Dating the genetic bottleneck of the African cheetah [J].Proc Natl Acad Sci USA, 90: 3172-3176.
Nei M. 1987. Molecular Evolutionary Genetics [M]. New York: Colombia University Press.
Okumura N, Kurosawa Y, Kobayashi E, Watanabe T, Ishiguro N, Yasue H, Mitsuhashi T. 2001. Genetic relationship amongst the major non-coding regions of mitochondrial DNAs in wild boars and several breeds of domesticated pigs [J].Anim Genet,32: 139-147.
Pestano J, Brown RP, Rodriguez F, Moreno A. 2000. Mitochondrial DNA control region diversity in the endangered blue chaffinch,Fringilla teydea[J].Mol Ecol,9: 1421-1425.
Pichler FB, Baker CS. 2000. Loss of genetic diversity in the endemic Hector’s dolphin due to fisheries-related mortality [J].Proc R Soc Lond:B Biol Sci,267: 97-102.
Rozas F, Rozas R. 1999. DnaSP version 3.0: an integrated program for molecular population genetics and molecular evolution analysis [J].Bioinformatics15: 174-175.
Schneider S, Roessli D, Excoffier L. 2000. Arlequin: A Software for Population Genetics Data Analysis: User manual ver 2000 [M]. Geneva: Genetics and Biometry Laboratory of Anthropology,University of Geneva.
Takeda K, Onishi A, Ishida N, Kawakami K, Komatsu M, Inumaru S. 1995. SSCP analysis of pig mitochondrial DNA D-loop region polymorphism [J].Anim Genet,26: 321-326.
Ursing BM, Arnason U. 1998. The complete mitochondrial DNA sequence of the pig (Sus scrofa) [J].J Mol Evol,47: 302-306.
Wallace DC. 1995. Mitochondrial DNA variation in human evolution, degenerative disease and aging [J].Am J Hum Genet57: 201-223.
Watanabe T, Hayashi Y, Kimura J, Yasuda Y, Saitou N, Tomita T, Ogasawara N. 1986. Pig mitochondrial DNA polymorphism, restriction map orientation and sequence data [J].Biochem Genet,24: 385-396.
Watanabe T, Ishiguro N, Okumura N, Nakano M, Matsui A, Hongo H, Ushiro H. 2001. Ancient mitochondrial DNA reveals the origin ofSus scrofafrom Rebun Island, Japan [J].J Mol Evol,52: 281-289.
Watanabe T, Okumura N, Ishiguro N, Nakano M, Matsui A, Sahara M, Komatsu M. 1999. Genetic relationship and distribution of the Japanese wild boar (Sus scrofa leucomystax) and Ryukyu wild boar (Sus scrofa riukiuanus) analyzed by mitochondrial DNA [J].Mol Ecol,8: 1509-1512.
Yildiz MA, Camdeviren H, Ozbeyaz C. 2002. Genetic diversity and divergence of some Swiss brown cattle herds in Turkey [J].Biochem Genet,40: 203-207.
Zhang YP, Oliver AR. 1993. Mitochondrial DNA evolution in theArctoidea[J].Proc Natl Acad Sci USA,90: 9557-9561.
云南地方猪种随机群体与选育群的遗传差异
亐开兴1,2,3, 吴桂生1,4, 苟 潇5, 严达伟5, 连林生5, Mumtaz Baig6, 张亚平1,2,*
(1. 中国科学院昆明动物研究所 云南省畜禽分子生物学重点实验室, 分子进化与基因组多样性实验室, 云南 昆明 650223; 2. 云南大学 生物资源保护与利用重点实验室, 云南 昆明 650091; 3. 云南省草地动物科学研究院, 云南 昆明 650212; 4. 中国科学院植物研究所, 云南 昆明 650204; 5. 云南农业大学 动物科技学院, 云南 昆明 650201; 6. Department of Zoology, Govt Vidarbha Institute of Science and Humanities, Amravati 444604, India)
为估计随机群体与选育群间的遗传差异, 测定了云南地方猪种 102个个体 438 bp的线粒体 DNA D-loop片段, 涉及4个随机群体、1个保种群和1个核心群。检测的16个多态位点界定了20个单倍型, 撒坝猪核心群和版纳微型猪保种群的单倍型数量仅为3个和1个, 而4个随机群体的单倍型数量都在6个以上, 4个随机群体的单倍型多态度极显著高于这两个选育群(h=0.732对0.425和0.000, exact test,P≤0.0036)。结果发现, 选育会导致遗传多样性过低, 尽管在随机群体中有较高的遗传多样性, 基于此, 及早干预和合理的育种计划将会有效地阻止核心群和选育群的遗传多样性下降。否则, 选育甚至会在短短几年内使遗传多样性程度急剧下降, 表明育种、保种与遗传多样性保存之间存在着尖锐冲突。该文还讨论了几个群体的遗传关系。
云南猪种; 遗传多样性; 随机群体; 选育群
S828.8; Q347
A
0254-5853-(2011)03-0255-07All mitochondrial DNA (mtDNA) variations originate from mutations (Brown et al, 1979; Wallace, 1995), which makes it ideal for studying genetic diversity of wild and domestic animals. Over the years mtDNA has become an efficient marker for analyzing the genetic relationships and divergence of intra- and inter-mammalian populations (Brown et al, 1982; Watanabe et al, 1986,1999,2001; Disotell et al, 1992; Zhang & Oliver, 1993; Luikart et al, 2001). The genetic relationship between wild boar and the domestic pig was reported by comparing the mtDNA (Watanabe et al, 1986, 1999; Giuffra et al, 2000; Okumura et al, 2001). The complete mtDNA sequence of the pig (Sus scrofa) was published (Ursing & Arnason, 1998). Ancient pig mtDNA was amplified and sequenced to clarify its origin and transition (Watanabe et al, 2001). Some authors held that Chinese pigs were introduced to improve European pig breeds during the 18th and early 19th centuries (Brown et al, 1982; Jone, 1998; Giuffra et al, 2000; Kijas & Anderson, 2001; Kim et al, 2002). Recent finding revealed that pigs were domesticated in the Mekong region and in the middle and downstream regions of the Yangtze River in East Asia (Wu et al, 2007). Although genetic diversity is a vital index for species diversity and conservation, humans depend on crossbreeding to improve the productivity and adaptability of livestock. Due to its harsh climate, geographical condition, and inconvenient transport, several local pig breeds were formed in the Yunnan Province of China (Lian et al, 2003). To protect and utilize the local pig resources, breeders have tried selection and crossbreeding (Lian et al, 2003). However, whether genetic diversity in maternally inherited mtDNA affects the success or failure of selection and conservation after selective breeding for some generations remains unclear. Consequently, we analyzed 102 pig samples for the 438 bp mtDNA control region to study the genetic diversity in randomly and selectively bred pig breeds from Yunnan.
2010-07-13;接受日期:2011-02-16
亐开兴(1975-), 男, 云南祥云人, 助理研究员, 专业方向:动物遗传育种与繁殖。E-mail: kaixqu@yahoo.com.cn
10.3724/SP.J.1141.2011.03255
date: 2010-07-13; Accepted date: 2011-02-16
s: This work was supported by research grants of the National Basic Research Program of China (2007CB815700; 2006CB102100), Bureau of Science and Technology of Yunnan Province, and Natural Science Foundation of China (30621092).
*Corresponding author (通信作者), Email: zhangyp@public.km.yn.cn
We thank Dr LIU Jin-Xian and Mr. SHAO Zhi-Yong for the discussion and GOU Shi-Kang, WU Shi-Fang, ZHU Chun-Ling, and ZHANG Li for technical assistance.
book=261,ebook=245
猜你喜欢
杂志排行
Zoological Research的其它文章
- 白介素1β对大鼠皮层神经元钠电流的急性作用
- 尼罗罗非鱼Orexin前体基因的克隆、组织分布及其在摄食调控中的表达
- Effects of urethane on the response properties of visual cortical neurons in young adult and old cats
- Phylogenetic evaluation of the taxonomic status of Papilio maackii and P. syfanius (Lepidoptera: Papilionidae)
- 阿魏酸对脂多糖诱导的小鼠小胶质细胞炎性反应的抑制作用
- 棕颈钩嘴鹛分子系统发育及分类关系初探
