Fuel source shift or cost reduction:Context-dependent adaptation strategies in closely related Neodon fuscus and Lasiopodomys brandtii against hypoxia
2022-08-05XiuJuanLiCongCongQiaoBoJianChenMengYangLiPengChenMaoLinHuangChunXiaoChenYanLiuHanChengMengWanJiangLuYeShiZhenLongWang
Xiu-Juan Li ,Cong-Cong Qiao ,Bo-Jian Chen ,Meng-Yang Li ,Peng Chen ,Mao-Lin Huang ,Chun-Xiao Chen,Yan Liu,Han Cheng,Meng-Wan Jiang,Lu-Ye Shi,4,*,Zhen-Long Wang,*
1 School of Life Sciences, Zhengzhou University, Zhengzhou, Henan 450001, China
2 College of Environmental Science and Engineering, Tongji University, Shanghai 200092, China
3 Jiaxing-Tongji Environmental Research Institute, Jiaxing, Zhejiang 314051, China
4 State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming,Yunnan 650223, China
5 Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, Yunnan 650223, China
6 School of Artificial Intelligence and Big Data, Henan University of Technology, Zhengzhou, Henan 450001, China
ABSTRACT Oxygen is essential for most life forms.Insufficient oxygen supply can disrupt homeostasis and compromise survival,and hypoxia-induced cardiovascular failure is fatal in many animals,including humans.However,certain species have adapted and evolved to cope with hypoxic environments and are therefore good models for studying the regulatory mechanisms underlying responses to hypoxia.Here,we explored the physiological and molecular responses of the cardiovascular system in two closely related hypoxiaadapted species with different life histories,namely,Qinghai voles (Neodon fuscus) and Brandt’s voles(Lasiopodomys brandtii),under hypoxic (10% O2 for 48 h) and normoxic (20.9% O2 for 48 h) exposure.Kunming mice (Mus musculus) were used for comparison.Qinghai voles live in plateau areas under hypoxic conditions,whereas Brandt’s voles only experience periodic hypoxia.Histological and hematological analyses indicated a strong tolerance to hypoxia in both species,but significant cardiac tissue damage and increased blood circulation resistance in mice exposed to hypoxia.Comparative transcriptome analysis revealed enhanced oxygen transport efficiency as a coping mechanism against hypoxia in both N.fuscus and L.brandtii,but with some differences.Specifically,N.fuscus showed upregulated expression of genes related to accelerated cardiac contraction and angiogenesis,whereas L.brandtii showed significant up-regulation of erythropoiesis-related genes.Synchronized upregulation of hemoglobin synthesis-related genes was observed in both species.In addition,differences in cardiometabolic strategies against hypoxia were observed in the rodents.Notably,M.musculus relied on adenosine triphosphate (ATP)generation via fatty acid oxidation,whereas N.fuscus shifted energy production to glucose oxidation under hypoxic conditions and L.brandtii employed a conservative strategy involving down-regulation of fatty acid and glucose oxidation and a bradycardia phenotype.In conclusion,the cardiovascular systems of N.fuscus and L.brandtii have evolved different adaptation strategies to enhance oxygen transport capacity and conserve energy under hypoxia.Our findings suggest that the coping mechanisms underlying hypoxia tolerance in these closely related species are context dependent.
Keywords:Heart;Hypoxia; Lasiopodomys brandtii;Neodon fuscus;RNA sequencing
INTRODUCTION
Oxygen is essential for the survival of most life forms and plays an important role in cellular aerobic energy supply and whole-body homeostasis (López-Barneo et al.,2001;Majmundar et al.,2010;Thompson,2016).A decrease in ambient oxygen can impair various physiological processes and trigger pathological reactions,thereby limiting organism survival in hypoxic environments,such as found at high altitude (Shao et al.,2021;Suzuki et al.,2019).Tissue-level hypoxia is often fatal and epidemiological studies have revealed that hypoxia is a principal cause of cardiovascular and cerebrovascular diseases,which are common causes of human death in industrialized societies (Chen et al.,2019;Iacobas et al.,2008).Hypoxia can also stimulate the production of reactive oxygen species in tissues,leading to oxidative stress and even death in severe cases (Hermes-Lima &Zenteno-Savn,2002;Lau et al.,2019).Therefore,understanding the response mechanisms to hypoxia is of great significance for the protection of humans and other animals in hypoxic environments and for diagnosis of hypoxiarelated diseases (Zhang et al.,2020).
The heart is a highly oxygen-consumptive organ and provides oxygen and nutrients to the body by continuously boosting blood flow (Katano et al.,2019).Therefore,a sustainable and sufficient oxygen supply is essential for normal cardiac function (Huang et al.,2021;Pell et al.,2016).Hypoxia can cause heart rate alteration,cardiac output disorder,blood pressure elevation,myocardial dysfunction,irreversible myocardial cell apoptosis,and even death(Khodaee et al.,2016;Paparde et al.,2015).However,some animals that live in hypoxic environments have evolved morphological and physiological cardiovascular adaptations to cope with such conditions,such as larger hearts,higher mitochondrial surface density,and greater microvascular density (Pichon et al.,2013;Wang et al.,2021).In terms of cardiac metabolism,some species have evolved improved energy efficiency by enhancing carbohydrate metabolism(Duan et al.,2021;Schippers et al.,2012;Su et al.,2021).Thus,these animals are considered good models for studying the regulatory mechanisms involved in the cardiovascular system in response to hypoxia.
The Qinghai vole (Neodon fuscus) is an endemic species distributed in the alpine meadows,swamp grasslands,and shrub grasslands of the Qinghai-Xizang (Tibet) Plateau,at elevations of 3 700–4 800 m a.s.l.(Zhang et al.,2013,2018).This small rodent has evolved a unique set of adaptation strategies to cope with its hypoxic environment,including more efficient oxygen transport capacity and lower risk of thrombosis (Li et al.,2021b).Brandt’s vole (Lasiopodomys brandtii) is closely related toN.fuscus(with both previously belonging to the genusLasiopodomys) and is mainly distributed in the temperate grasslands of Inner Mongolia,southeastern Mongolia,and parts of southern Russia,at an average elevation of <1 000 m a.s.l.(Liu et al.,2020;Shi et al.,2021a).Qinghai voles live in a persistently hypoxic environment,whereas Brandt’s voles only experience relatively mild and periodic hypoxia when gathered underground during specific periods,such as at rest.However,empirical studies suggest thatL.brandtiialso exhibits extraordinary tolerance to hypoxia (Dong et al.,2018;Li et al.,2021c;Shi et al.,2021b,2021c).Although these two species are closely related,their different habitats and life history patterns may have contributed to differences in hypoxic adaptations.Therefore,in the present study,we examined whether the high hypoxia tolerance of these two closely related species is achieved through different coping mechanisms and strategies.We conducted comparative physiological and transcriptomics analyses to investigate changes in cardiac tissue structure,blood parameters,and mRNA expression inN.fuscus,L.brandtii,and Kunming mice(Mus musculus) under hypoxic (10% O2for 48 h) and normoxic (20.9% O2for 48 h) conditions.
MATERIALS AND METHODS
Ethics statement
All protocols used in the present study were approved by the Institutional Animal Care and Use Committee,Zhengzhou University,Zhengzhou,Henan,China (permit No.ZZUIRB2022-44),and were carried out in accordance with the approved guidelines for the Use of Experimental Animals(China).All local regulations and laws were strictly followed.
Animals and acclimation
TheN.fuscusvoles were captured in the Guoluo Tibetan Autonomous Prefecture,Qinghai Province,China (N34°8′,E100°11′).TheL.brandtiivoles were purchased from the Chinese Academy of Agricultural Sciences (Beijing,China).The Kunming mice was purchased from the Laboratory Animal Center of Henan Province (Zhengzhou,China).All experimental animals were raised individually in uniformly sized polycarbonate boxes (length×width×height,290 mm×178 mm×160 mm).Air temperature was 20–24 °C,relative humidity was 35%–50%,and photoperiod was 12 h light:12 h dark.Animals were fed mixed commercial rat feed and rabbit feed (Henan Experimental Animal Center,Zhengzhou,Henan Province,China) as well as fresh carrots and waterad libitum.All experimental animals were domesticated in the laboratory for at least eight weeks.
Hypoxia treatment
After acclimation,18 adult females of similar size were selected from each species and randomly divided into two groups:hypoxia treatment group (10% O2for 48 h,n=9) and normoxia treatment group (20.9% O2for 48 h,n=9).Hypoxic treatment was conducted in December using hypoxic chambers (DS-II hyperbaric cabin,Huaxin Hyperbaric Cabin,China),with the O2level monitored in real time using an oxygen analyzer.A bottle containing sodium hydroxide was placed in the cabin to absorb the carbon dioxide released by the animals.Animals in the normoxia treatment group were also placed in a sealed oxygen chamber for 48 h.Except for the oxygen content,all other environmental conditions were constant for all groups.No mortality occurred during hypoxia/normoxia treatment.After hypoxia/normoxia treatment,all experimental animals were euthanized immediately by an intraperitoneal overdose injection of pentobarbital sodium (30 mg/kg) and whole hearts were removed for further analysis.Histological and hematological analyses were performed in six of the nine animals of each species in each group and transcriptome sequencing after RNA extraction was conducted in the remaining three animals from each group.
Histological evaluation
Whole hearts were collected from the animals and fixed in 4%paraformaldehyde for more than 24 h,dehydrated with ethanol,embedded in paraffin,and sectioned into 4 μm thick sections,which were processed and stained with hematoxylin and eosin (H&E) and Masson trichrome to evaluate tissue damage.H&E-stained sections of the cardiac apex of normoxic and hypoxic groups were examined under a light microscope (BX53 fluorescence microscope,Olympus Corporation,Japan) at 200× magnification.Masson staining was performed to assess the degree of cardiac fibrosis in five randomly selected visual fields (400× magnification) in each sample by calculating the percentage of blue-stained area(corresponding to fibrosis) relative to total heart area using ImageJ v1.53 (Wang et al.,2018).GraphPad Prism v8.0.1(GraphPad Software,USA) was used to analyze and graph differences in cardiac fibrosis among the six samples from the normoxic and hypoxic treatment groups with independent samplet-test.Data are presented as mean±standard error of the mean (SEM) andP<0.05 was considered statistically significant.
Measurement of hematological parameters
Fresh blood samples from the three species were collected in ethylenediamine tetra-acetic acid (EDTA)-anticoagulated tubes to measure erythrocyte system parameters using an automated hematology analyzer (BC-2800Vet,Mindray,China),including red blood cells (RBC),hemoglobin (HGB),hematocrit (HCT),mean corpuscular volume (MCV),mean red blood cell hemoglobin (MCH),mean red blood cell hemoglobin concentration (MCHC),and red blood cell distribution width(RDW).GraphPad Prism v8.0.1 was used to analyze and graph the differences between the normoxic and hypoxic treatment groups using independent samplet-test.Data are presented as mean±SEMandP<0.05 was considered statistically significant.
RNA extraction,cDNA library construction,and RNA sequencing (RNA-seq)
Total RNA was extracted from heart tissue using TRIzol reagent (Invitrogen,USA) according to the manufacturer’s instructions.RNA quality was assessed by:(1) analysis of RNA integrity by agarose gel electrophoresis (1.0%);(2)measurement of RNA concentration and purity using a NanoDropND-1000 spectrophotometer (NanoDrop Technologies,LLC,USA);and (3) accurate detection of RNA integrity number (RIN) using an Agilent 2100 bioanalyzer(Agilent Technologies,USA).RNA samples meeting these standards were added with oligo (dT) magnetic beads to enrich mRNA with poly-A tails,which were then broken into short fragments (250–300 bp) using fragmentation buffer and used as a template to synthesize first-and second-strand cDNA.The obtained double-stranded cDNA was then purified using AMPure XP beads and subjected to end-repair,poly-A tail addition,sequencing adaptor ligation,fragment size selection,and polymerase chain reaction (PCR) amplification for cDNA library construction.Finally,the Illumina NovaSeq 6000 platform was used to perform next-generation pairedend sequencing of the constructed cDNA library,with 150 paired-end reads and >6 Gb of raw data per sample.The remaining RNA samples were reverse-transcribed into cDNA using a PrimeScript RT Reagent Kit and gDNA Eraser(TaKaRa,Japan) according to the manufacturer’s instructions and stored at −80 °C for subsequent quantitative real-time PCR (qRT-PCR).
De novo assembly and quality assessment
Fastp v0.20.1 (Chen et al.,2018) was used to filter reads containing adapters and low-quality reads (reads containingN>0.002 or >50% bases with aQ-value≤20) to acquire clean data to ensure the quality and reliability of data analysis.The clean reads were then assembled into transcripts using Trinity v2.8.5 (Grabherr et al.,2011;Haas et al.,2013) and clustered into unigenes with Corset v1.09 (Davidson &Oshlack,2014)for hierarchical clustering by removing transcript redundancy and selecting the longest transcripts as unigenes.The quality of the assembled transcripts and unigenes was assessed by evaluating three criteria:i.e.,gene coverage,assembly statistics,and ortholog completeness.Gene coverage was calculated by comparing the clean reads from each sample ofN.fuscus,L.brandtii,andM.musculuswith their assembled transcripts or unigenes.Assembly statistics of transcripts and unigenes of the three species were checked using TrinityStats.pl script,including total number,total assembled bases,average contig length,and N50 and GC content.Ortholog completeness of unigenes was examined using BUSCO v4.0.5 (Simão et al.,2015) against a mammalian database with transcriptome mode.The length distribution of the unigenes was calculated using Perl scripts,which were uploaded to GitHub (https://github.com/lxjbio/Calculate-lengthdistribution).
Functional annotation of unigenes
The functions of the unigenes were examined by searching the homologous sequences of all unigenes against six public databases,including the National Center for Biotechnology Information (NCBI) nonredundant database (Nr),Swiss-Prot database,euKaryotic Orthologous Groups (KOG),eggNOG orthology data (EggNOG),Kyoto Encyclopedia of Genes and Genomes (KEGG),and Gene Ontology (GO) database forN.fuscus,L.brandtii,andM.musculus.For Nr,Swiss-Prot,and KOG annotations,the BLASTX program (BLAST+suite,v2.9.0) was used to identify putative functions for homologs with an E-value cutoff of 1e−5,with the best hit retained.For EggNOG annotation,eggnog-mapper v2.0.1 (Huerta-Cepas et al.,2017) was used to identify orthologs with default parameters based on the eggNOG orthology v5.0 database(Huerta-Cepas et al.,2019).For KEGG annotation,KEGG Automatic Annotation Server (KAAS,https://www.genome.jp/kegg/kaas/) was retrieved to obtain KEGG orthology assignments and pathways using the singledirectional best-hit method (Batista et al.,2019;Moriya et al.,2007;Zeng et al.,2019).For GO annotation,Blast2GO(Conesa et al.,2005) implemented in OmicsBox v1.3.3(https://www.biobam.com/omicsbox/) was used to classify unigenes into three categories (biological process,cellular component,and molecular function) to provide a dynamically updated standardized vocabulary for assigning the functions of genes and their products based on the top 20 BLAST results from Swiss-Prot.The distribution of GO function classification for the unigenes was then plotted using OmicShare (https://www.omicshare.com/tools).
Analysis of differential gene expression
Gene expression levels were calculated by mapping the clean reads ofN.fuscus,L.brandtii,andM.musculusto their respective unigenes using bowtie2 in the RSEM package (Li &Dewey,2011).The obtained count value was exported to DESeq2 (Love et al.,2014) for differential expression analysis between the hypoxic and normoxic treatment groups and the resultingP-values were adjusted using the Benjamini and Hochberg approach to control for the false discovery rate(FDR).Unigenes with |log2(fold-change)|≥1 and adjustedP<0.05 were designated as differentially expressed genes(DEGs).Volcano and heatmap plots of DEGs were drawn using the ggplot2 package and R software,respectively.
Functional enrichment and protein-protein interaction(PPI) analysis
Information regarding DEG functions was collected from unigene annotations and the DEGs were examined using GO and KEGG enrichment analyses to determine potential biological functions and metabolic pathways.The GO and KEGG enrichment analyses were performed using hypergeometric statistical tests,andP-values were corrected for multiple testing.GO terms and KEGG pathways with enrichment atP<0.05 (modified Fisher’s exact test) and more than two genes enriched were considered as significantly enriched functions (Koga et al.,2021;Lavergne et al.,2020;Liu et al.,2015).Hub genes were identified by constructing PPI networks using the Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org/)with the Markov clustering (MCL) algorithm,and subsequently visualized in Cytoscape v3.8.2 (Shannon et al.,2003).
qRT-PCR analysis and validation
Six genes,including two oxygen transport-associated genes and four randomly selected genes,were chosen for qRT-PCR to verify the reliability of our RNA-seq data and analysis.Primers for qRT-PCR were designed using Oligo7 software and further confirmed online using the Primer-BLAST tool available at NCBI (https://www.ncbi.nlm.nih.gov/tools/primerblast/).The qRT-PCR procedure was performed in a 25 μL reaction mixture (12.5 μL of UltraSYBR mixture,0.5 μL of PCR forward primer (10 μmol/L),0.5 μL of PCR reverse primer (10 μmol/L),1 μL of template cDNA,and 10.5 μL of ddH2O) using a LightCycler 480 (Roche Diagnostics,Germany) under the following conditions:polymerase activation at 95 °C for 10 min,followed by 40 cycles of DNA denaturation at 95 °C for 10 s,primer annealing temperature for 30 s,extension at 72 °C for 32 s,and final melt-curve analysis at 95 °C for 15 s,60 °C for 1 min,95 °C for 15 s,and 60 °C for 15 s to measure nonspecific PCR products.All qRTPCR analyses were performed using three biological repeats and three technical repeats to minimize experimental error,and the whole process was performed on ice and in darkness to avoid degradation of experimental samples.Relative gene expression levels were calculated using the 2−ΔΔCtmethod with β-actin as the internal control (Kashyap et al.,2015;Xu et al.,2019).GraphPad Prism v8.0.1 was used to analyze the relationship between gene expression levels measured by qRT-PCR and RNA-seq using Pearson correlation analysis.
RESULTS
Histological changes in the heart
H&E staining revealed that heart morphology inN.fuscusandL.brandtiidid not change significantly in response to hypoxia,whereas the intercellular spacing between myocardial cells inM.musculusincreased significantly (Figure 1A–F).Masson staining revealed that collagen fiber content in theN.fuscusandL.brandtiihearts did not change significantly after exposure to hypoxia but increased significantly inM.musculus(P<0.01;Figure 1G;Supplementary Figure S1).
Hematological parameters
Among the seven hematological indicators tested,N.fuscusshowed a significantly lower MCV after hypoxia treatment compared with normoxia treatment (P<0.05;Figure 1H).Mice showed significantly higher HCT,MCV,and RDW after hypoxia treatment (P<0.05;Figure 1J),whereasL.brandtiishowed no significant differences in any index in response to hypoxia (Figure 1I).
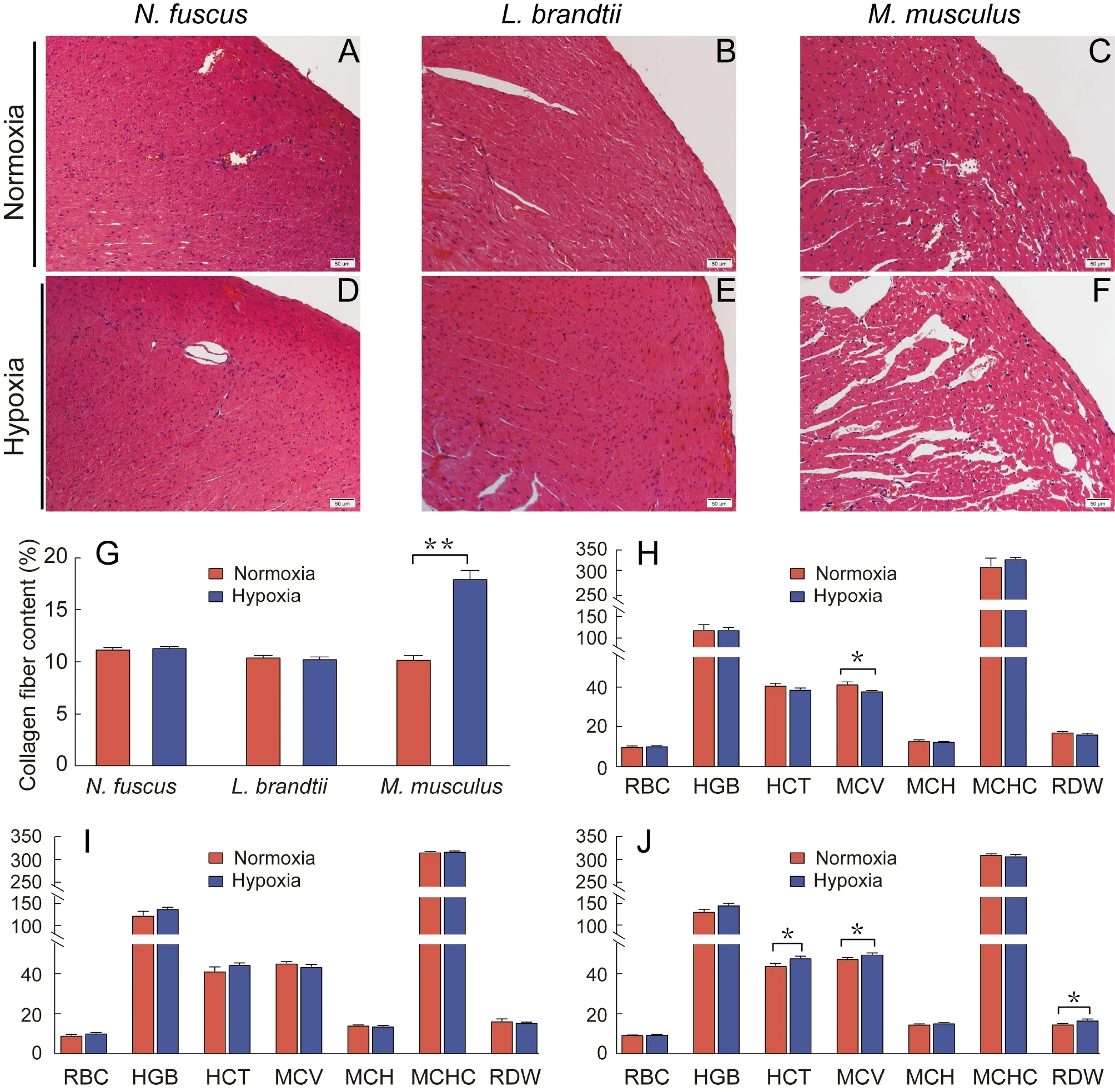
Figure 1 Changes in heart tissue morphology,collagen fiber content,and erythrocyte system before and after hypoxia treatment
RNA-seq and assembly
The RNA quality of all samples was assessed using a cutoff RIN value ≥7.5,which showed that the quality of the RNA samples was good (Supplementary Table S1).RNA-seq yielded a total number of 1 035 084 256 paired reads forN.fuscus,L.brandtii,andM.musculus(Supplementary Table S2).After removing the adaptors and filtering low-quality reads,1 018 238 884 clean reads were retained,with >95.05%Q30 bases in each sample (Supplementary Table S2).De novoassembly generated 528 549 transcripts (mean length,858.35 bp) hierarchically clustered into 153 192 unigenes(mean length,1 102.67 bp) inN.fuscuswith an N50 (minimum contig length required to cover 50% of unigenes) value of 1 748 bp (Table 1).The same method was used to obtain 471 494 transcripts (mean length,881.75 bp) and 132 299 unigenes (mean length,1 150.20 bp) with an N50 value of 1 915 bp forL.brandtii,and 471 657 transcripts (mean length,864.98 bp) and 137 036 unigenes (mean length,1 112.32 bp)with an N50 value of 1 847 bp forM.musculus(Table 1).Moreover,the filtered clean reads from each sample were aligned to the transcripts and unigenes with a mapping rate of 92.16% and 87.09%,respectively.The completeness of the unigenes was close to 80% (Table 1) and the main length distribution range of the unigenes ranged from 500 to 1 000 bp in the three species (Supplementary Figure S2A and Table S3).
Functional annotation and classification of unigenes
InN.fuscus,L.brandtii,andM.musculus,44 404 (28.99%),41 384 (31.28%),and 45 967 (33.54%) unigenes,respectively,were annotated in at least one of the six databases(Supplementary Table S4),and 15 589,15 701,and 15 140 unigenes,respectively,were assigned to all six databases(Supplementary Figure S2B–D).ForN.fuscus,L.brandtii,andM.musculus,Nr annotation identified 39 110 (25.53%),37 302(28.20%),and 41 306 (30.14%) unigenes,respectively,with homologous proteins (Supplementary Figure S2 and Table S4).Swiss-Prot annotation identified 33 455 (21.84%),32 025(24.21%),and 33 001 (24.08%) unigenes,respectively,with homologous proteins (Supplementary Figure S2 and Table S4).For KOG analysis,24 113 (15.74%),23 124 (17.48%),and 23 144 (16.89%) unigenes,respectively,were assigned to 25 functional classifications (Supplementary Figures S2,S3and Table S4).For GO annotation,28 280 (18.46%),27 838(21.04%),and 27 480 (20.05%) unigenes,respectively,were assigned to one or more terms of the three categories(Supplementary Figures S2,S4 and Table S4).
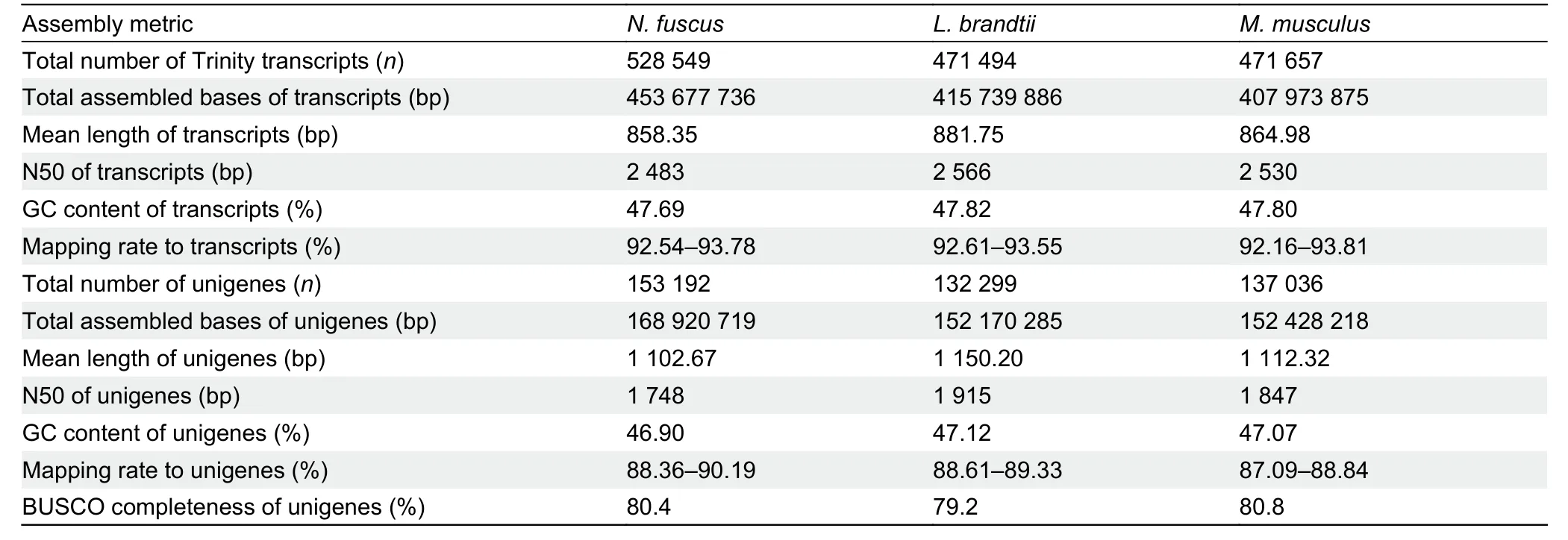
Table 1 Transcriptome assembly metrics and assessment results in N.fuscus,L.brandtii,and M.musculus
DEG identification
Comparative transcriptome analysis was performed between the normoxia and hypoxia treatment groups to identify DEGs in response to hypoxic stress.As a result,788 DEGs were identified inN.fuscus,including 286 up-regulated and 502 down-regulated genes (Supplementary Figure S5A),among which,71 up-regulated and 119 down-regulated genes were annotated (Supplementary Tables S5,S6).Likewise,572 DEGs were identified inL.brandtii,including 267 up-regulated and 305 down-regulated genes (Supplementary Figure S5B),among which 75 up-regulated and 117 down-regulated genes were annotated (Supplementary Tables S5,S6).In addition,1 297 DEGs were identified inM.musculus,including 831 upregulated and 466 down-regulated genes (Supplementary Figure S5C),among which 244 up-regulated and 136 downregulated genes were annotated (Supplementary Tables S5,S6).Furthermore,cluster analysis of the DEGs showed that the three biological replicates in the normoxia and hypoxia treatment groups were clustered together for each species(Supplementary Figure S5D–F).
Functional enrichment analysis of DEGs
InN.fuscus,the up-regulated DEGs were significantly enriched in 120 GO terms,including 112 biological process terms,five cellular component terms,and three molecular function terms (Figure 2A;Supplementary Table S7).These enriched terms were mainly related to cellular processes(single-organism cellular process,transport,establishment of localization,cell differentiation,localization,cell morphogenesis,cellular developmental process,cellular component morphogenesis,and cell morphogenesis involved in differentiation),development (blood vessel morphogenesis,blood vessel development,and vasculature development),and signal transduction (negative regulation of signal transduction,negative regulation of signaling,positive regulation of signal transduction,positive regulation of signaling,regulation of signal transduction,and regulation of signaling).The down-regulated DEGs were only significantly enriched in seven GO molecular function terms (Figure 2B;Supplementary Table S7),which were all related to binding activity (ion binding,nucleic acid binding transcription factor activity,transcription factor activity,sequence-specific DNA binding,protein binding,bridging,binding,bridging,lipid binding,and DNA binding).
InL.brandtii,the up-regulated DEGs were significantly enriched in 31 GO terms,including 24 biological process terms,five cellular component terms,and two molecular function terms (Figure 3A;Supplementary Table S7).Most enriched GO terms were related to stimulus and regulation(response to stimulus,regulation of biological quality,homeostatic process,and biological regulation),cellular processes (transport,establishment of localization,localization,plasma membrane organization,endomembrane system organization,single-organism membrane organization,membrane,cytoskeleton organization,multicellular organismal process,single-organism cellular process,cell differentiation,intracellular,and cytoplasm),development (single-organism developmental process,developmental process,and anatomical structure development),and enzyme activity(oxidoreductase activity).The down-regulated DEGs were significantly enriched in 42 GO terms,including 20 biological process terms,13 cellular component terms,and nine molecular function terms (Figure 3B;Supplementary Table S7).The enriched terms were mainly related to metabolic process (small molecule metabolic process,single-organism metabolic process,lipid metabolic process,catabolic process,cellular amino acid metabolic process,organic acid metabolic process,carboxylic acid metabolic process,oxoacid metabolic process,secondary metabolic process,nitrogen cycle metabolic process,and cofactor metabolic process,and other terms) and enzyme activity (mitochondrion,oxidoreductase activity,catalytic activity,transferase activity,lyase activity,isomerase activity,and enzyme regulator activity).

Figure 2 GO enrichment,KEGG enrichment,and PPI results for up-regulated and down-regulated DEGs in N.fuscus
InM.musculus,the up-regulated DEGs were significantly enriched in 28 GO terms,including 15 biological process terms,nine cellular component terms,and four molecular function terms (Figure 4A;Supplementary Table S7).Most enriched GO terms were related to stimulus and regulation response of stimulation (response to stress,response to stress,and response to stimulus),metabolic process(metabolic process,cellular protein metabolic process,protein metabolic process,and sulfur compound metabolic process),cellular processes (cellular macromolecular complex assembly,cell adhesion,and aging),and enzyme activity(isomerase activity and enzyme regulator activity).The downregulated DEGs were significantly enriched in 12 GO terms,including eight biological process terms,two cellular component terms,and two molecular function terms(Figure 4B;Supplementary Table S7).These enriched terms were mainly related to cell process (cell proliferation,multicellular organismal process,single-multicellular organism process,protein maturation,aging,and extracellular region),development (embryo development,multicellular organism development,and single-organism developmental process),and enzyme activity (enzyme regulator activity).
Metabolic pathway enrichment analysis of DEGs
InN.fuscus,the up-regulated DEGs were significantly enriched in 21 pathways related to cellular processes(regulation of actin cytoskeleton,mitophagy-animal,and tight junction),environmental information processing (sphingolipid signaling pathway and vascular endothelial growth factor(VEGF) signaling pathway),human disease (hepatitis C,choline metabolism in cancer,salmonella infection,human cytomegalovirus infection,pathogenicEscherichia coliinfection,shigellosis,African trypanosomiasis,and malaria),metabolism (arachidonic acid metabolism and linoleic acid metabolism),and organismal systems (osteoclast differentiation,serotonergic synapse,axon guidance,chemokine signaling pathway,platelet activation,and leukocyte transendothelial migration) (Figure 2C;Supplementary Table S8).The down-regulated DEGs were significantly enriched in 10 pathways related to cellular processes (P53 signaling pathway),environmental information processing (MAPK signaling pathway),human disease(proteoglycans in cancer,endocrine resistance,basal cell carcinoma,human papillomavirus infection,bladder cancer,and transcriptional misregulation in cancers),metabolism(glyoxylate and dicarboxylate metabolism),and organismal systems (estrogen signaling pathway) (Figure 2D;Supplementary Table S8).

Figure 3 GO enrichment,KEGG enrichment,and PPI results for up-regulated and down-regulated DEGs in L.brandtii
InL.brandtii,the up-regulated DEGs were significantly enriched in six pathways related to cellular processes(apoptosis),human disease (herpes simplex infection,platinum drug resistance,African trypanosomiasis,and malaria),and metabolism (glycine,serine,and threonine metabolism) (Figure 3C;Supplementary Table S8).The downregulated DEGs were significantly enriched in 31 pathways related to human disease (platinum drug resistance and chemical carcinogenesis),metabolism (carbon metabolism,tyrosine metabolism,glutathione metabolism,tryptophan metabolism,nitrogen metabolism,glycine,serine,and threonine metabolism,ubiquinone and other terpenoidquinone biosynthesis,arachidonic acid metabolism,porphyrin and chlorophyll metabolism,pentose and glucuronate interconversions,phenylalanine,tyrosine,and tryptophan biosynthesis,arginine biosynthesis,cysteine and methionine metabolism,ascorbate and aldarate metabolism,phenylalanine metabolism,metabolic pathways,linoleic acid metabolism,drug metabolism-cytochrome P450,drug metabolism-other enzymes,biosynthesis of amino acids,metabolism of xenobiotics by cytochrome P450,retinol metabolism,and steroid hormone biosynthesis),and organismal systems (PPAR signaling pathway,thyroid hormone synthesis,serotonergic synapse,bile secretion,cholesterol metabolism,and complement and coagulation cascades) (Figure 3D;Supplementary Table S8).
InM.musculus,the up-regulated DEGs were significantly enriched in 14 pathways related to cellular processes(autophagy-animal,p53 signaling pathway,and autophagyother eukaryotes),environmental information processing(apelin signaling pathway and Jak-STAT signaling pathway),human disease (pancreatic cancer,platinum drug resistance,and human cytomegalovirus infection),metabolism(propanoate metabolism,tyrosine metabolism,valine,leucine,and isoleucine degradation,metabolic pathways,and nicotinate and nicotinamide metabolism),and organismal systems (PPAR signaling pathway) (Figure 4C;Supplementary Table S8).The down-regulated DEGs were significantly enriched in 14 pathways related to cellular processes (focal adhesion,adherens junction,tight junction,and apoptosis),environmental information processing (hippo signaling pathway),human disease (bladder cancer,microRNAs in cancer,thyroid cancer,proteoglycans in cancer,and pathogenicEscherichia coliinfection),metabolism(glycosphingolipid biosynthesis-lacto and neolacto series),and organismal systems (oxytocin signaling pathway,phototransduction-fly,and thyroid hormone signaling pathway)(Figure 4D;Supplementary Table S8).

Figure 4 GO enrichment,KEGG enrichment,and PPI results for up-regulated and down-regulated DEGs in M.musculus
Validation of RNA-seq results by qRT-PCR
All qRT-PCR experiments were performed using specific primers for amplification of target genes (Supplementary Table S9).The expression profiles of six genes,includingHbaandAlas2,which are involved in oxygen transport,showed a similar trend (Figure 5A–C) and were highly significantly correlated (R2=0.9676,P<0.01 forN.fuscus,R2=0.9802,P<0.01 forL.brandtii,andR2=0.9581,P<0.01 forM.musculus;Figure 5D–F) to the RNA-seq assay,indicating the reliability of our transcriptome analysis.

Figure 5 Gene expression levels measured by qRT-PCR and RNA-seq methods
DISCUSSION
Hypoxia can disrupt homeostasis and lead to impaired cardiac function and even death in many organisms,including humans.Studying the hypoxic response mechanism of the cardiovascular system in animals that have adapted to such environments can provide valuable information for the protection of humans and other animals.In the present study,we assessed the response and regulation of the cardiovascular system to hypoxia in two closely related rodents,N.fuscusandL.brandtii,and usedM.musculusfor comparison.Compared with the mice,N.fuscusandL.brandtiiexhibited a strong tolerance to hypoxia,but via different adaptation mechanisms.
Physiological response to hypoxia
Histological analysis revealed that the cardiac tissue morphology and collagen fiber content ofN.fuscusandL.brandtiidid not change significantly in response to hypoxia compared with normoxia (Figure 1;Supplementary Figure S1),indicating a strong tissue-level tolerance to hypoxia.However,inM.musculus,myocardial cell spacing and tissue fibrosis increased significantly,indicating that external hypoxia caused tissue damage (Figure 1;Supplementary Figure S1).Hematological analysis revealed that MCV was significantly reduced inN.fuscusunder hypoxic conditions,which may help decrease blood viscosity,reduce blood circulation resistance,and facilitate oxygen diffusion in hypoxic environments (Kong et al.,2019;Verghese,2013).In contrast,MCV and HCT were significantly increased inM.musculus,consistent with previous study (Liu et al.,2010;Roklicer et al.,2020),which may increase blood circulation resistance.In addition,RDW was also significantly increased inM.musculus,which can rapidly increase in response to RBC rupture.Notably,increased RDW is significantly negatively correlated with RBC deformation and may increase cardiac load and cardiovascular disease (Yoon et al.,2015).The elevation of these three blood parameters inM.musculusmay be due to hypoxia promoting erythropoietin production,thereby inducing the formation of enlarged RBCs (Yčas et al.,2015).
Transcriptome response to hypoxia
Enhanced oxygen transport efficiency in N.fuscus and L.brandtii under hypoxia:Results showed thatN.fuscusandL.brandtiicoped with hypoxia by enhancing oxygen transport efficiency,and this functional convergence appears to be due to varied gene regulation patterns.Myh6andMyh7,two key genes encoding αMyHC and βMyHC proteins,respectively,were significantly up-regulated in theN.fuscusheart under hypoxic conditions (Figures 2E,6;Supplementary Table S6).High expression of these genes in mammals is associated with accelerated cardiac contraction and economical contractility (Grimes et al.,2017;Miyata et al.,2000;Yin et al.,2015).Therefore,we speculate that the significant increase inMyh6andMyh7may enhance cardiac contractility inN.fuscusin hypoxic conditions and improve oxygen transport efficiency.In addition to the up-regulation of genes involved in cardiac contraction,genes related to angiogenesis and endothelial cell proliferation were also significantly up-regulated (Rac1,Pla2g4c,Alox15,Nrp1,Cxcl12,Rock2,andNotch4) or downregulated (Krit1,Thbs1,andAngptl7) inN.fuscusunder hypoxic conditions,with most genes enriched in the VEGF signaling pathway,blood vessel morphogenesis,blood vessel development,vasculature development,and other pathways(Figures 2,6;Supplementary Table S6).Among these genes,Rac1plays an important role in regulating the proliferation and migration of vascular smooth muscle cells and the rearrangement of endothelial cells (Sawada et al.,2010).Significant up-regulation ofPla2g4candAlox15promotes the release of arachidonic acid,which,in turn,promotes prostaglandin production,thereby promoting angiogenesis(Barnett et al.,2010;Singh &Rao,2019).Significant upregulation ofNrp1,Cxcl12,Rock2,andNotch4may also have promoted angiogenesis and vascular development inN.fuscusto some extent (He et al.,2010;Kryczek et al.,2005;Mehta et al.,2018;Trindade &Duarte,2020).In agreement,we also found significant down-regulation inKrit1,Thbs1,andAngptl7,which encode antiangiogenic proteins (Figures 2E,6;Supplementary Table S6) and negatively regulate angiogenesis (Lawler,2002;Orso et al.,2013;Toyono et al.,2015).Based on these findings,we speculate thatN.fuscusmay promote angiogenesisin vivovia a complex network of multiple genes,thereby enhancing blood circulation and facilitating oxygen transport under hypoxic conditions.
Spta1,Slc4a1,andEpb42were significantly up-regulated inL.brandtiiunder hypoxic conditions and strongly interacted withHbaandHbb(Figure 3E;Supplementary Table S10).Of note,these three genes encode spectrin,band 3,and protein 4.2,respectively,which are important skeletal proteins in the erythrocyte membrane (Cox et al.,2019;Long et al.,2015;Schneider et al.,2009;Wang et al.,2017a;Wu et al.,2019).Therefore,the synergistic and significant up-regulation ofSpta1,Slc4a1,andEpb42may helpL.brandtiimaintain stable erythrocyte morphologyin vivounder hypoxic conditions to ensure oxygen supply (Figure 6).Band 3 also acts as a transporter to mediate electron neutralization and anion exchange in the cell membrane,promoting the exchange of chloride and bicarbonate on the erythrocyte plasma membrane and the elimination of carbon dioxide from tissues(Arakawa et al.,2015).Bpgm,which encodes an allosteric regulator of hemoglobin,was up-regulated in response to hypoxia (Supplementary Table S6).This gene promotes the release of oxygen by regulating the affinity of O2and hemoglobin when blood flows through tissues (Sun et al.,2016).
In addition to different regulatory mechanisms,we also identified synchronized gene expression regulation shared byN.fuscusandL.brandtii.For example,the expression levels ofHbaandHbb,which encode the α-globin and β-globin subunits for hemoglobin,respectively,were significantly upregulated in both species (Figures 2E,3E,6 and Supplementary Table S6).Hemoglobin is the main respiratory protein in mammalian RBCs and plays a central role in binding and transporting oxygen to support the energy and metabolic requirements essential for life (Zhang et al.,2015).Species living in hypoxic environments for prolonged periods have shown an increase in the ability of hemoglobin to absorb and transport oxygen by up-regulating the expression ofHbaandHbbin vivo(Dong et al.,2018;Yang et al.,2018).Here,PPI analysis showed a strong synergistic up-regulation inAlas2,Hba,andHbbin bothN.fuscusandL.brandtii(Figures 2E,3E;Supplementary Table S10).Alas2,which encodes 5-aminolevulinic acid synthase 2 in erythroid cells,synthesizes heme for hemoglobin (Abu-Farha et al.,2005) and its upregulation increases heme content in RBCs (Hofer et al.,2003;Khechaduri et al.,2013).Heme binds to globin subunits encoded byHbaandHbbto form hemoglobin subunits,with the two hemoglobin α and two hemoglobin β subunits then forming adult hemoglobin with heterotetramers to transport oxygen (Brown et al.,2016;Ludwig et al.,2016).Therefore,we speculate thatN.fuscusandL.brandtiimaintain sufficient oxygen supply to various tissues and organs under hypoxic conditions via the coordinated regulation ofHba,Hbb,andAlas2.
Mus musculusmice do not reside in hypoxic environments and lack effective oxygen transport strategies to respond to hypoxia.In terms of myocardial contraction,Acta1andActc1,which encode alpha-cardiac actin and alpha-skeletal actin,respectively,were significantly down-regulated (Figure 4E;Supplementary Table S6).These two genes are closely linked to myocardial contraction and cardiomyocyte apoptosis(Bildyug,2019;Frade et al.,2013;Ilkovski et al.,2005).NppaandCorinwere also significantly up-regulated inM.musculusunder hypoxic conditions and strongly interacted withActa1andActc1(Figure 4E;Supplementary Table S6).Increased expression ofNppaandCorinis significantly correlated with physiological and pathological reactions,such as hypoxia,hypertension,myocardial ischemia,ventricular hypertrophy,and heart failure (Xu-Cai &Wu,2010;Yan et al.,2000).These findings are contrary to the enhanced myocardial contraction characteristics found inN.fuscusunder hypoxic conditions,thus highlighting the differences in tolerance to hypoxia in these two species.We also identified a series of angiogenesis-related genes (Nrp2,Snail2,Ets1,Plau,andVash1) that interacted withVegfaand were significantly downregulated inM.musculusin response to hypoxia (Figure 4E;Supplementary Table S6),which is not conducive to the formation of blood vessels (Boosani and Sudhakar,2011;Cheng et al.,2019;Lv et al.,2014;Sindi et al.,2020;Zheng et al.,2013).

Figure 6 Dynamic deduction of oxygen transport regulation in N.fuscus and L.brandtii under hypoxic conditions

Figure 7 Cardiac energy metabolism pathways activated under hypoxic environments
Context-dependent cardiometabolic strategies inN.fuscus andL.brandtii under hypoxia:InN.fuscus,cardiac metabolism under hypoxic conditions underwent a metabolic shift from fatty acid to glucose oxidation (Figure 7A),with 11 down-regulated genes significantly enriched in lipid binding(Figure 2B;Supplementary Table S7).Among these downregulated genes,Acot11encodes acyl-CoA thioesterases,which catalyze the hydrolysis of long-chain fatty acyl-CoAs to form free fatty acids.Down-regulation of this gene may reduce fatty acid synthesis (Diessler et al.,2018;Jeckel et al.,2014).The down-regulation ofAcss2was significantly enriched in ion binding and glyoxylate and dicarboxylate metabolism(Supplementary Tables S7,S8).Acss2plays an important role in catalyzing acetyl-CoA production during fatty acid oxidation and a significant down-regulation of this gene can result in a significant reduction in acetyl-CoA (Martínez-Micaelo et al.,2016;Sun et al.,2018).In contrast to the significant downregulation of genes related to fatty acid oxidation,we observed a significant up-regulation inAldob,a core gene related to glucose oxidation,inN.fuscusunder hypoxic conditions (Figure 7A).Aldobencodes aldolase B (also known as fructose-bisphosphate aldolase B),a key glycolytic enzyme in vertebrates that catalyzes the conversion of fructose 1,6-bisphosphate to glyceraldehyde 3-phosphate,which is further catabolized to pyruvate (Miller et al.,2012;Pamidimukkala et al.,2018;Potter et al.,2021).Pyruvate is converted into acetyl CoA,which drives the tricarboxylic acid (TCA) cycle and thus energy production via oxidative phosphorylation (Kurniawan et al.,2020).Heme is an indispensable prosthetic group of complex III and complex IV and increased synthesis can promote oxidative phosphorylation (Brennan et al.,2014;Shinzawa-Itoh et al.,2016).
In the adult mammalian heart,more than 70% of adenosine triphosphate (ATP) is produced by oxidation of fatty acids and the remaining is generated by carbohydrates (Hsiao et al.,2021;Peoples et al.,2019).However,under conditions of normal oxygen consumption,carbohydrates (glucose) can produce more ATP than fatty acids,as glucose has higher oxidation efficiency (Bai et al.,2021;Lorenzo et al.,2013).The conversion of animal cardiometabolic substrates from fatty acids to glucose under hypoxic conditions is considered an oxygen-saving strategy,which can improve cardiometabolic efficiency and maintain ATP production(Abdurrachim et al.,2015;Duan et al.,2021;Su et al.,2021).In high-altitude Andean mice,cardiac metabolism promotes glucose oxidation by increasing the activity of hexokinase as a strategy to conserve oxygen compared with low-altitude counterparts (Schippers et al.,2012).Similarly,based on transcriptome analysis,genes involved in glycolysis are upregulated in Tibetan pigs compared with Yorkshire pigs,suggesting that glycolysis may be more active in Tibetan pigs to reduce oxygen demand and maintain energy production(Duan et al.,2021).Furthermore,31P magnetic resonance spectroscopy of high-altitude humans (Sherpas) reveals metabolic organization of the heart to preferentially use carbohydrates (glucose) instead of fatty acids (Hochachka et al.,1996).Even though shifting the “fuel source” from fatty acid to glucose oxidation may be advantageous under hypoxia,it also has disadvantages,such as cardiac hypertrophy (Kolwicz et al.,2012;Ma et al.,2019).Hence,increasing the ratio of glucose oxidative energy in heart tissue may be an important adaptation strategy employed byN.fuscusliving in hypoxic conditions to maintain an efficient energy supply.
UnlikeN.fuscus,L.brandtiireduced both fatty acid and glucose oxidation in the heart under hypoxic conditions(Figure 7B).In terms of glucose metabolism,Aldobexpression,which was significantly up-regulated inN.fuscus,was significantly down-regulated inL.brandtii,thereby inhibiting the conversion of fructose 1,6-bisphosphate to glyceraldehyde 3-phosphate.Pkm,which encodes pyruvate kinase,was also significantly down-regulated (Figure 7B;Supplementary Table S6),thus inhibiting the production of pyruvate and ATP from phosphoenolpyruvate (Fan et al.,2019).Changes in fatty acid oxidation included the significant down-regulation of two key genes,Acox1andEhhadh,which catalyze the conversion of acyl-CoA to 3-ketoacyl-CoA(Figure 7B;Supplementary Table S6),thus inhibiting fatty acid oxidation inL.brandtii(Chen et al.,2016;Qu et al.,2019).Interestingly,under hypoxic conditions,theL.brandtiihearts showed significant down-regulation ofPopdc2(Supplementary Table S6),which is highly expressed in cardiac pacing and conduction systems and plays an important regulatory role in heart rate dynamics (Froese et al.,2012;Milanesi et al.,2015).No or low expression ofPopdc2in the heart can cause severe arrhythmia and sinus arrest symptoms,which can reduce the average heart rate and lead to a bradycardia phenotype (Froese et al.,2012;Schindler &Brand,2016).Bradycardia and insufficient cardiac energy supply may affect exercise performance inL.brandtiiunder hypoxic environments,with previous studies showing that nerve synapse excitability and athletic ability are significantly reduced inL.brandtiiunder hypoxic conditions (Dong et al.,2018).Previous studies have also reported that low heart and metabolic rates are common strategies to conserve oxygen in hypoxia-tolerant species (Arieli et al.,1986;Grimes et al.,2017;Kirby et al.,2018;Pamenter et al.,2018).Lasiopodomys brandtiivoles face hypoxic conditions when resting in their tunnels,with low-level exercise and energy requirements.Therefore,we speculate that appropriate downregulation of heart rate and energy supply may be an adaptive mechanism to conserve energy consumption under hypoxic conditions (Grimes et al.,2014;Kirby et al.,2018).
In contrast toN.fuscusandL.brandtii,theM.musculushearts did not exhibit an enhanced glucose oxidation energy supply ratio or a decreased heart rate in relation to reduced energy consumption under hypoxic conditions,and cardiac energy supply was mainly dependent on fatty acid oxidation,consistent with its main source of energy in normoxic conditions (Figure 7C).The mice showed significant upregulation of four fatty acid oxidation-related genes,includingAcer2,Acsl1,Acss1,andAcadm,which are involved in catalyzing ceramides,long-chain fatty acids,and short-chain fatty acids to produce intermediate products in the betaoxidation of fatty acids (Chen et al.,2014;Li et al.,2018;Paul et al.,2014;Wang et al.,2017b).Nevertheless,a single,inflexible energy supply and inadequate substrate oxygen may result in an insufficient cardiac energy supply inM.musculusunder hypoxia,thereby limiting its survival under these conditions.
Based on the above,our findings suggest that the two vole species exhibit differences in oxygen transport and energy utilization strategies in response to hypoxia,which is,to some extent,corroborated by previous studies in other animals.For example,bar-headed geese cope with the hypobaric and hypoxic conditions encountered when flying over the Himalayas by increasing cardiac output (Scott et al.,2015),while other animals that live in hypoxic environments,such as lizards,naked mole rats,and blind mole rats,possess stronger cardiovascular system-related oxygen transport capacity (He et al.,2013;Li et al.,2021a).Thus,combined with our findings on voles,we suggest that enhanced oxygen transport capacity may be a hypoxic adaptation strategy shared by species living in hypoxic environments.In terms of energy utilization,both the energy transformation strategy ofN.fuscus(i.e.,fatty acid oxidation to glucose oxidation) and oxygen-saving strategy ofL.brandtii(i.e.,reducing both fatty acid and glucose oxidation) under hypoxic environments have been confirmed laterally in other studies (Duan et al.,2021;Hochachka et al.,1996;Ivy et al.,2020;Schippers et al.,2012).
CONCLUSIONS
In the present study,we investigated the physiological and molecular responses of the cardiovascular system of two closely related rodents,N.fuscusandL.brandtii,under hypoxia (10% O2for 48 h) and normoxia (20.9% O2for 48 h),withM.musculusused for comparison.Results showed enhanced oxygen transport efficiency as a coping mechanism against hypoxia inN.fuscusandL.brandtii,but with some differences.Specifically,N.fuscusshowed up-regulation of genes related to accelerated cardiac contraction and angiogenesis,whereasL.brandtiishowed significant upregulation of erythropoiesis-related genes.The synchronized up-regulation of hemoglobin synthesis-related genes was also observed in both species.We also found differences in cardiometabolic strategies against hypoxia.Mus musculusrelied on ATP generation via fatty acid oxidation,whereasN.fuscusshifted energy production to glucose oxidation under hypoxic conditions andL.brandtiiemployed a conservative strategy involving down-regulation of fatty acid and glucose oxidation and a bradycardia phenotype.These strategies may helpN.fuscusandL.brandtiiconserve energy and cope with hypoxia.Our findings suggest that the coping mechanisms of these closely related and hypoxia-tolerant species are context dependent.
DATA AVAILABILITY
The raw sequencing data were deposited in the NCBI under BioprojectID PRJNA787778 and in GSA under accession No.CRA006487 and in Science Data Bank under DOI:10.11922/sciencedb.01649.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
L.Y.S.conceived and designed the study.X.J.L.and L.Y.S.analyzed the data,prepared the figures,and wrote the first manuscript draft.C.C.Q.,B.J.C.,and Z.L.W.participated in the revision of the manuscript.M.Y.L.,P.C.,M.L.H.,C.X.C.,Y.L.,H.C.,and M.W.J.were responsible for animal collection and husbandry.All authors read and approved the final version of the manuscript.
杂志排行
Zoological Research的其它文章
- Zoological Research call for papers of Cavefish Special Issue
- Ecological study of cave nectar bats reveals low risk of direct transmission of bat viruses to humans
- Population and conservation status of a transboundary group of black snub-nosed monkeys (Rhinopithecus strykeri) between China and Myanmar
- Nucleus accumbens-linked executive control networks mediating reversal learning in tree shrew brain
- Europe vs.China:Pholcus (Araneae,Pholcidae) from Yanshan-Taihang Mountains confirms uneven distribution of spiders in Eurasia
- Author Correction:A new species of the gudgeon genus Microphysogobio Mori,1934 (Cypriniformes:Cyprinidae) from Zhejiang Province,China
