花生晚斑病抗病育种研究进展
2019-12-09吴丽军李双铃韩锁义夏晗迟玉成任艳石延茂尹亮王兴军袁美
吴丽军 李双铃 韩锁义 夏晗 迟玉成 任艳 石延茂 尹亮 王兴军 袁美
摘要:花生晚斑病是一种世界性病害,可导致花生减产和品质降低,防治该病害最经济有效的方法是抗病品种的应用。本文介绍了花生晚斑病的危害和防治途径,对花生晚斑病的抗性种质资源挖掘、抗性遗传及抗病基因、分子标记、抗病品种的培育等多个方面的最新研究进展进行了综述,同时对加强花生晚斑病研究提出了一些建议。
关键词:花生;晚斑病;病害防治;抗病育种
中图分类号:S565.203.4 文献标识号:A 文章编号:1001-4942(2019)09-0177-08
Advances in Peanut Breeding for Resistance to Late Leaf Spot
Wu Lijun Li Shuangling Han Suoyi2, Xia Han3, Chi Yucheng Ren Yan Shi Yanmao
Yin Liang Wang Xingjun3, Yuan Mei1
(1.Shandong Peanut Research Institute/Key Laboratory of Peanut Biology and Genetic Improvement, Ministry of Agriculture,
Qingdao 266100, China; 2. Industrial Crops Research Institute, Henan Academy of Agricultural Sciences, Zhengzhou 450002,
China; 3.Biotechnology Research Center, Shandong Academy of Agricultural Sciences, Jinan 250100, China)
Abstract Late leaf spot (LLS) disease is one of the most important leaf diseases in peanut (Arachis hypogaea L.), which could cause substantial yield losses and reduce the seed quality. Application of resistant varieties is one of the most economical and effective methods for controlling this disease. In this paper, the damage and controlling ways of LLS were summarized. The recent research progress on identification of resistant germplasm resources, resistance inheritance and disease resistant genes, molecular markers, breeding of disease-resistant varieties and so on were reviewed. Moreover, some suggestions were put forward for strengthening LLS research.
Keywords Peanut; Late leaf spot; Disease management; Disease resistant breeding
花生是世界上重要的經济和油料作物之一,在食用植物油及休闲食品中占有举足轻重的地位。花生叶斑病在所有花生产区均有发生或流行,是造成花生减产的重要因素之一。花生叶斑病包括褐斑病和黑斑病。褐斑病又叫“早斑病”(early leaf spot),黑斑病又称“晚斑病”(late leaf spot),其中晚斑病是花生生产中最常见、危害最大的病害之一[1]。花生晚斑病主要危害叶片,破坏叶绿素,造成光合作用效能下降,大量病斑造成叶片枯死、脱落,严重影响干物质积累和荚果成熟。在印度,晚斑病流行较重[2];我国北方花生产区以晚斑病流行为主,南方产区早斑病和晚斑病常交替发生,多数年份以晚斑病危害为主[3-5]。花生晚斑病可使花生减产10% ~ 20%,严重的可达50%以上,每年对全球花生生产造成的损失达6亿美元[ 6-9]。
1 花生晚斑病病原菌
花生晚斑病病原菌为短胖孢[Cercosporidium personatum(Beck & Curtis) Deighton],无性阶段为半知菌类短胖孢属落花生短胖孢,有性阶段为子囊菌纲球腔菌属伯克利球腔菌(Mycosphaerella berkeleyi)。晚斑病菌侵染后,花生叶正面产生较小的黑褐色、圆形或近圆形病斑,背面通常产生许多黑色小点,即病菌分生孢子梗和分生孢子。晚斑病病菌主要以分生孢子座和菌丝体在病株残体上越冬,或以分生孢子黏附在荚果、茎秆表面越冬。分生孢子萌生芽管直接从花生叶片表皮或气孔侵入。22 ~ 23℃时,3 ~ 4 d显病,1周开始产孢。翌年借风雨传播进行初侵染和再侵染[5,10]。
2 花生晚斑病的主要防治途径
花生晚斑病主要防治途径有:杀菌剂的应用、生物防治、综合管理措施等[5]。杀菌剂的推广应用是当前田间防治晚斑病的重要措施,但对花生叶斑病的防治效果仅在60%左右[10],且化学农药的不合理施用会引起人畜中毒、环境破坏和植物药害等一系列环境、社会问题。目前,生物防治措施的研究报道较少,直接应用于花生大规模生产实践中的更少。综合管理措施主要通过创造不利于晚斑病发病的微生态环境或在发病前期尽早预报预防晚斑病的发生发展。培育和利用抗病品种是经济、安全且最有效的防控病害的途径。在同样病原菌压力的情况下,抗病品种比感病品种发病慢、感病轻。抗病品种的应用可以减少杀菌剂的使用、降低生产成本、减轻环境污染压力和由于农药残留导致的食品安全风险。
3 花生晚斑病抗病种质资源
抗病种质资源,即抗源,是植物抗病育种的原始材料,也是选育优良抗病品种的遗传物质基础,系统地搜集、保存、评价和研究抗源是抗病育种研究最重要的基础工作。种质资源主要包括地方品种、改良品种、近缘植物和通过种内杂交、远缘杂交、诱发变异获得的抗病中间材料等。各国科学家一直致力于发掘花生晚斑病的抗病种质资源。
3.1 花生野生种抗晚斑病种质资源的筛选
花生不同种质间抗、感病程度存在一定差异,尤其是花生野生种存在着优异的抗病基因。与栽培种相比,野生种花生具有较高的遗传多样性,能够适应一系列复杂环境,是抵抗生物胁迫和非生物胁迫的重要基因来源[11-13]。梁炫强等从34份花生野生种中发掘出24份高抗晚斑病材料,1份中抗材料[14]。Abdou等对南美洲和非洲不同地区花生的不同亚种、野生变种和栽培品种进行抗病鉴定,发现Arachis cardenasii对晚斑病免疫[15]。花生区组的A. duranensis、A. spegazzini、A. correntina、A. stenosperma、A. diogoi(=A. chacoense),直立区组的A. paraguriensis,三籽粒区组的A. pussila,围脉区组的A. villosulicarpa,根茎区组的A. hagenbeckii、A. glabrata、A. burkartii,大根区组的A. repens等野生种以及一些未定名的野生材料对晚斑病均表现出高抗或免疫[16]。
3.2 花生栽培种抗晚斑病种质的筛选与创制
国内外花生育种工作者致力于抗晚斑病种质资源的筛选与创制,筛选获得的抗性种质部分已经应用到花生育种实践中。Hassan等从16个不同种质材料中筛选出可作为叶斑病抗性育种最有优势的亲本品种NC 3033、NC 5和AC 3139[17]。Kornegay等研究发现NC 3033和NC-GP 343在筛选的6个花生品系中抗性最高,是花生晚斑病抗性育种最具潜力的品系[18]。Cook从12份花生栽培种中鉴定出PI 259747和PI 341879高抗晚斑病[19]。1986年,Chiteka等从116个花生基因型中检测到对晚斑病抗性最强的基因型为UF81206-1、UF81206-2.72 × 32b-3-2-2-2-2-2-l-b3-b和US 29-b3-B(85701)[20]。Anderson等从500份引进花生种质筛选出33份对晚斑病表现部分抗性[21]。Dwivedi等从15份种间杂交材料中筛选出ICGV 9006、99013、99004、9903和99001共5个晚斑病抗病育种亲本材料[22]。2001年,国际半干旱研究所(ICRISAT)鉴定发现ICGV# 99001和99004种质系抗晚斑病[23]。Chapin等评价了47个弗吉尼亚型育种系和8个品种对晚斑病的抗性,发现栽培种Bailey、三个姊妹系(N03088T、N03089T和N03090T)和N03091T始终不容易被晚斑病侵染[24]。美国的Southern runner属于中抗晚斑病品种[25],玻利维亚地方品种Bayo grande以及其他一系列的育种系也具有较好的叶斑病抗性[26],澳大利亚新品种Sutherland对晚斑病表现高抗[27],ICG11337在抗晚斑病育种中是一个很好的抗性供体[28]。
我国科研工作者对国外引种的花生种质和国内育成品种也开展了叶斑病抗性鉴定。黎穗临从981份国外花生资源中鉴定出抗叶斑病品种722份,其中高抗276份,高抗早斑病和晚斑病的品种144份,同时从333份广东省花生资源中筛选鉴定出57份抗叶斑病品种,如遂溪勾鼻、顶子细粒等[29]。董炜博等研究发现引进品种UF91108对晚斑病高抗,可作为花生育种的抗源材料[30]。袁虹霞等筛选出对花生早斑病和晚斑病抗性较好的开农31、濮花8030和豫花15号等系列品种[31]。
4 花生晚斑病的抗性遗传、QTL定位及分
子标记与抗性相关基因[BT)]
4.1 花生晚斑病的抗性遗传
花生晚斑病抗性遗传较为复杂,有研究表明叶斑病抗性由2对以上核隐性基因控制,而且早斑病和晚斑病抗性遗传是分别独立的[3, 32]。Nevill在前人研究的基础上进一步得出花生晚斑病抗性是由5对隐性基因控制[33]。Soriano等发现花生叶斑病突变体的抗性由2个突变基因IS-1和IS-2控制,這2个突变基因编码的蛋白主要通过抑制病原菌的生长从而对病原菌产生抗性抑制[34]。Pasupuleti等研究表明花生栽培种与野生种种间杂交衍生系ICG11317和ICG13919对晚斑病的抗性受细胞核基因和细胞质基因的共同控制[28]。夏友霖等的研究认为ICGV86699对花生晚斑病的抗性受 2 对加性-上位性主基因 + 加性-上位性多基因控制[35]。
4.2 花生晚斑病的QTL定位及分子标记
近年来,花生晚斑病QTL及分子标记陆续被筛选、鉴定。早期主要以AFLP分子标记和SSR分子标记为主。2007年,夏友霖等以中花5号 × ICGV 86699的F2群体为材料,利用AFLP分析结合BSA法,鉴定出与晚斑病抗性紧密连锁的三个AFLP标记:E35/M51、E37/M48和E41/M47[36]。Leal-Bertioli等以二倍体野生种间杂交的F2群体为材料鉴定出5个晚斑病抗性QTLs和34个序列特定的候选基因片段[37]。Khedikar等以来自杂交组合TAG24 × GPBD4的268份RIL群体为材料,利用SSR标记构建了14个遗传连锁群,鉴定出11个晚斑病抗性QTLs,可解释晚斑病1.70%~6.50%的表型变异[38]。Sujay等利用分别来自两个组合TAG24 × GPBD和TG 26 × GPBD4的RIL群体,鉴定出15个晚斑病抗性QTLs[39]。Shoba等利用TMV 2 × COG 0437的F2群体和SSR标记技术,筛选出与晚斑病抗性相关的SSR标记PM 384-100[40]。Wang等利用SSR分子标记技术及组合Tifrunner × GT-C20构建的群体,在F2图谱(5.3 cM/位点)中鉴定出54个QTLs,其中37个为叶斑病QTLs,在F5图谱(5.7 cM/位点)中鉴定出23个QTLs,其中13个为叶斑病QTLs[41]。Pandey等利用相同的RIL群体共鉴定出42个QTLs,其中抗早斑病9个,抗晚斑病22个[42]。Khera等利用SunOleic 97R和NC94022的重组自交系群体,构建了包含248个标记位点的群体遗传连锁图谱,鉴定出48个QTLs,其中22个抗早斑病QTLs,20个抗晚斑病QTLs,并发现共有6个主要的基因组区域包含的QTLs控制不止一种抗病性[43]。
随着测序技术的进步及测序成本的降低,SNP标记用于花生分子遗传图谱的构建和QTL定位。2014年,Zhou等利用简化基因组测序技术构建了来自中花5号 × ICGV 86699组合的RIL群体高密度遗传图[44],鉴定出20个晚斑病抗性QTLs,并发现2个QTLs(qLLSB6-7和qLLSB1)位于编码NB-LRR的两个基因簇中[45]。Pandey等利用基于全基因组重测序(WGRS)的QTL-seq方法分析来自RIL群体(TAG24 × GPBD)的抗池/感池,鉴定出A03上的一个2. 98 Mb的基因组区(131.67 ~ 134.65 Mb)同时负责锈病和晚斑病抗性,开发了1个抗晚斑病SNP诊断标记[46]。Clevenger等也利用QTL-seq方法对来自组合Florida-07 × GP-NC WS的重组自交系群体的抗池/感池开展测序分析,鉴定出3个分别位于A05、B05和B03染色体上花生晚斑病QTLs[47];Chu等利用花生58 K SNP芯片数据分析该群体,构建了一个基于SNP的包含855个位点的连锁图谱,鉴定出三个晚斑病抗性QTLs(qLLSA05、qLLSB03和qLLSB05)[48],其中qLLSA05、qLLSB03与用QTL-seq策略[47]获得的相同。Liang等利用品种Tamrun OL07和高抗叶斑病育种系Tx964117构建的F2∶6重组自交系(RIL)群体,共鉴定出6个与花生叶斑病相关的QTLs[49]。Lu等通过构建一个包含5 874个位点的遗传图谱,在A05连锁群约0.38 cM的区域发现了一个抗晚斑病的主效QTL,该区域包含26个候选基因,其中一些被注释与其它物种的抗病性调节有关[50]。Han等利用分布于20个连锁群的2 753个SNP标记构建了一张标记平均间距为1.34 cM的高分辨率图谱,将一个主效晚斑病抗性QTL定位在B05染色体上[51]。Agarwal等利用重组自交系群体的WGRS数据,建立基于SNP的高密度遗传图谱,共鉴定出35个主要效应QTLs,其中2个晚斑病QTLs分别在A05(PVE 47.63%)和B03(34.03%)上,并开发验证了相关KASP标记[52]。这些鉴定出的QTLs、分子标记和抗病基因丰富的基因组区域将有助于花生抗病基因的克隆和分子标记辅助抗病品种的培育。
4.3 花生晚斑病抗性相关基因
关于花生晚斑病抗性相关基因的研究近年有较多报道。Luo等通过分析不同抗性花生品种在接种晚斑病菌后384个转录本的变化情况,寻找抗性基因片段,结果检测到代表56个功能类别基因的112个基因上调表达,并验证了其中的17个基因[53]。二倍体野生种A. diogoi接种晚斑病菌48 h后部分cDNA上调表达,其中一个编码类环膦素蛋白基因AdCyp,该基因受病原菌和植物激素的上调表达影响,转AdCyp的烟草对青枯菌的抗性增强、对寄生疫霉菌的感病性降低[54]。Guimares等对接种晚斑病菌后的野生种进行转录组学分析,获得多个响应晚斑病侵染的差异表达基因,并通过qRT-PCR对5个抗性基因类似物和4个逆转录转座子序列进行了验证,同时开发了多个特异性抗性分子标记[55]。病原菌相关蛋白PR-5和防御素属于植物抗真菌蛋白,转SniOLP(Solanum nigrum osmotin-like protein,龙葵渗调蛋白)和RsAFP2(Raphanus sativus antifungal protein-2,萝卜抗真菌蛋白-2)双基因的花生对晚斑病的抗性明显增强[56]。另外,来自水稻和烟草的几丁质酶基因、芥菜生物防御素基因转化到花生后均能提高花生对叶斑病的抗性[57-59]。
5 抗花生晚斑病品种的培育
常规抗病品种选育的主要途径包括引种、系统选种、常规杂交育种、远缘杂交育种和诱变育种等。研究表明,高抗病可以与产量、品质因素相结合,其中一些品系可能在生产中发挥很好的作用[60]。美国农业部农业研究服务部和佐治亚大学农业与环境科学院联合选育的花生新品种Tifrunner,对早、晚斑病均具有中等抗性[61],FloridaMDR98、C-99R和York等均为抗叶斑病品种[23, 62]。佐治亚大学海岸平原实验站将PI 203396和品种AgraTech GK 7雜交育成了高抗早、晚斑病的Georganic[63]。
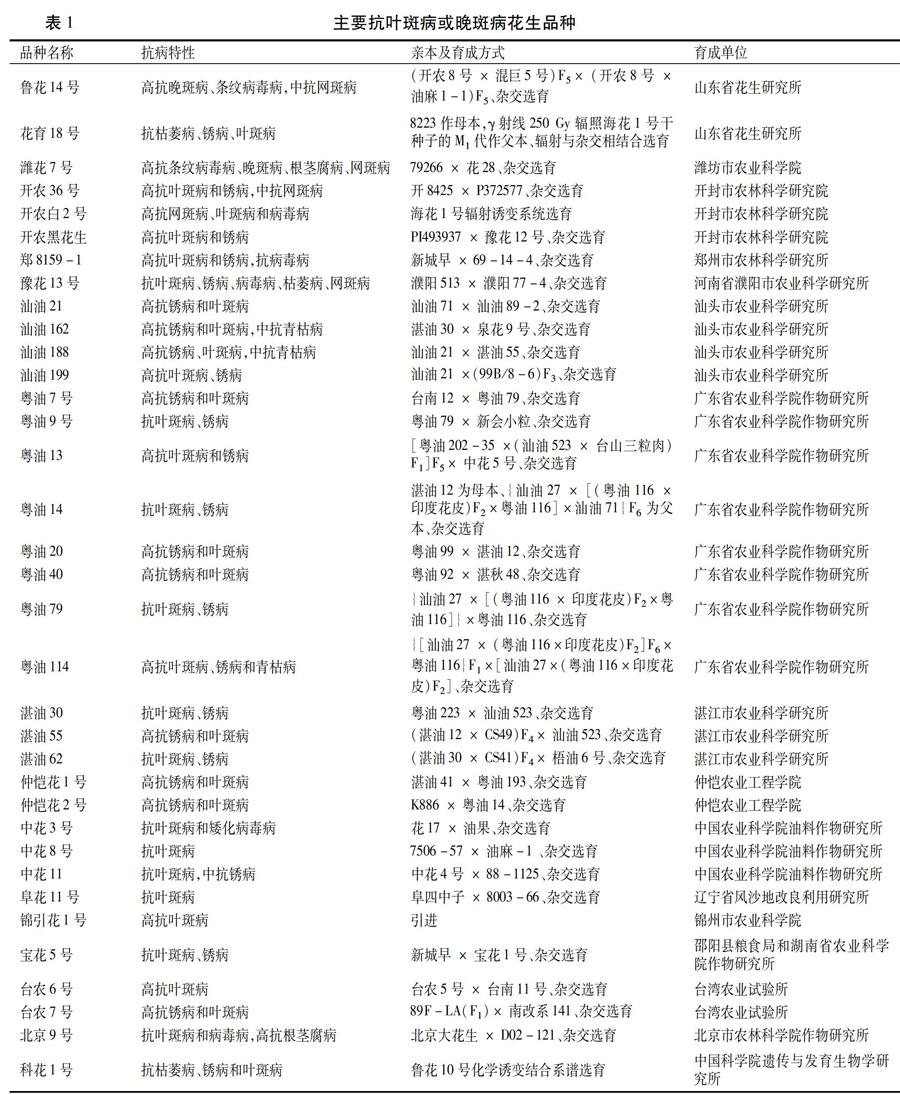
从20世纪90年代开始,我国开展了大量花生晚斑病抗病育种研究,并育成了一些抗或耐晚斑病的花生品种[16],对叶斑病或晚斑病的抗性品种进行汇总,见表1。山东省主要推广花生品种有72个,其中抗晚斑病或叶斑病的有3个;河南省主要推广品种有61个,其中抗晚斑病或叶斑病的有5个;广东省主要推广品种有42个,其中抗晚斑病或叶斑病的有17个;湖北省主要推广品种有22个,其中抗晚斑病或叶斑病的有3个;辽宁省主要推广品种有45个,其中抗晚斑病或叶斑病的有2个;湖南省主要推广品种有11个,其中抗晚斑病或叶斑病的有1个;台湾主要推广品种约有22个,其中抗叶斑病的有2个;北京市主要推广品种约有23个,其中抗晚斑病或叶斑病的有2 个。
6 存在的问题与展望
花生是我国重要的油料作物之一,在保障我国食用油供给方面具有重要作用。晚斑病防治是花生生产的重要环节之一。运用分子标记技术可以缩短育种年限,将多个抗性基因聚合到同一个材料中,已经成为花生育种的一个重要方向。全面阐明花生晚斑病抗性遗传机制,并根据生产需要定向培育综合性状优良、高抗花生晚斑病的新品种是花生科研工作者面临的重要课题之一。
目前,对花生晚斑病抗病机理和抗病育种的研究相对较少,例如,花生晚斑病抗病机理目前仍未阐明;花生抗晚斑病的分子标记开发虽然取得了一定成效,但在品种选育中真正发挥作用的极少,花生抗晚斑病的育种主要还是以传统的常规育种方式为主;多数育成的抗病品种还是依赖栽培种之间的杂交选育,很多野生种质材料对多种病害高抗甚至免疫,但由于倍性差异和杂交亲和性低等原因,野生资源的利用还非常有限。因此,对花生晚斑病应加强以下几个方面的研究:(1)结合栽培花生和野生花生全基因组测序的研究成果,通过比较基因组学,鉴定晚斑病抗性关键基因,深入研究花生晚斑病抗病分子机理;(2)充分挖掘抗病花生种质的遗传多样性,将分子标记辅助选择与常规育种相结合,聚合多个抗性基因,开展多抗优质花生新品种的培育;(3)通过基因工程改良、种间杂交、人工四倍体的合成等途径,将野生资源中的抗病基因渐渗到栽培种中,创造大量栽野渐渗系,为晚斑病抗性育种奠定基础。
参 考 文 献:
[1]Miller I L, Norden A J, Knauft D A, et al. Influence of maturity and fruit yield on susceptibility of peanut to Cercosporidium personatum(late leaf spot pathogen)[J].Peanut Science, 1990, 17(2): 52-58.
[2]Mcdonald D, Subrahmanyam P, Gibbons R W, et al. Early and late leaf spots of groundnut[M].International Crops Research Institute for the Semi-Arid Tropics, 1985.
[3]趙守贤. 花生的抗病育种[J].花生科技, 1993(2): 25-27.
[4]沈一, 刘永惠, 陈志德. 花生叶斑病研究概述[J].花生学报, 2014, 43(2): 42-46.
[5]韩锁义, 张新友, 朱军, 等. 花生叶斑病研究进展[J].植物保护, 2016, 42(2): 14-18.
[6]万书波. 中国花生栽培学[M].上海: 上海科学技术出版社, 2003.
[7]Zhang S, Reddy M S, Kokalis-Burelle N, et al. Lack of induced systemic resistance in peanut to late leaf spot disease by plant growth-promoting rhizobacteria and chemical elicitors[J].Plant Disease, 200 85(8): 879-884.
[8]Dwivedi S L, Crouch J H, Nigam S N, et al. Molecular breeding of groundnut for enhanced productivity and food security in the semi-arid tropics: opportunities and challenges[J].Advances in Agronomy, 2003, 80(4): 153-221.
[9]Ogwulumba S I, Ugwuoke K I, Iloba C. Prophylactic effect of paw-paw leaf and bitter leaf extracts on the incidence of foliar myco-pathogens of groundnut (Arachis hypogaea L.) in Ishiagu, Nigeria[J].African Journal of Biotechnology, 2008, 7(16): 2878-2880.
[10]鄢洪海, 张茹琴. 环境因子对花生黑斑病菌生长发育的影响[J].湖北农业科学, 2009, 48(9): 2145-2148.
[11]Mallikarjuna N, Jadhav D R, Reddy K, et al. Screening new Arachis amphidiploids, and autotetraploids for resistance to late leaf spot by detached leaf technique[J].European Journal of Plant Pathology, 2012, 132(1): 17-21.
[12]朱琳, 靳秋生, 郭凤丹, 等. 花生野生种在花生抗病中的利用[J].中国油料作物学报, 2019, 41(3): 469-477.
[13]Singh A K, Mehan V N S. Sources of resistance to groundnut fungal and bacterial diseases: an update and appraisal[M].International Crops Research Institute for the Semi-Arid Tropics, 1997.
[14]梁炫强, 黎穗临, 叶维霖, 等. 国外花生种质资源的引进、鉴定和利用[J].花生科技, 1999(S1): 130-135.
[15]Abdou Y A, Gregory W C, Cooper W E. Sources and nature of resistance to Cercospora arachidicola Hori and Cercosporidium personatum (Beck & Curtis) Deighton in Arachis species[J].Peanut Science, 1974, 1(1): 6-11.
[16]禹山林. 中国花生品种及其系谱[M].上海: 上海科学技术出版社, 2008.
[17]Hassan H N, Beute M. Evaluation of resistance to Cercospora leaf spot in peanut germplasm potentially useful in a breeding program[J].Peanut Science, 1977, 4(2): 78-83.
[18]Kornegay J L, Beute M K, Wynne J C. Inheritance of resistance to Cercospora arachidicola and Cercosporidium personatum in six Virginia-type peanut lines[J].Peanut Science, 1980, 7(1): 4-9.
[19]Cook M. Susceptibility of peanut leaves to Cercosporidium personatum[J].Phytopathology, 198 71(8): 787-791.
[20]Chiteka Z A, Gorbet D W, Shokes F M, et al. Components of resistance to late leafspot in peanut. Ⅰ. levels and variability-implications for selection[J].Peanut Science, 1988, 15(1): 25-30.
[21]Anderson W F, Holbrook C C, Brenneman T B. Resistance to Cercosporidium personatum within peanut germplasm[J].Peanut Science, 1993, 20(1): 53-57.
[22]Dwivedi S L, Pande S, Rao J N, et al. Components of resistance to late leaf spot and rust among interspecific derivatives and their significance in a foliar disease resistance breeding in groundnut (Arachis hypogaea L.)[J].Euphytica, 2002, 125(1): 81-88.
[23]Singh A K, Dwivedi S L, Pande S, et al. Registration of rust and late leaf spot resistant peanut germplasm lines[J].Crop Science, 2003, 43(1): 440-442.
[24]Chapin J W, Thomas J S, Isleib T G, et al. Field evaluation of Virginia-type peanut cultivars for resistance to tomato spotted wilt virus, late leaf spot, and stem rot[J].Peanut Science, 2010, 37(1): 63-69.
[25]Singh M P, Erickson J E, Boote K J, et al. Late leaf spot effects on growth, photosynthesis, and yield in peanut cultivars of differing resistance[J].Agronomy Journal, 201 103(1): 85-91.
[26]Gremillion S, Culbreath A, Gorbet D, et al. Response of progeny bred from Bolivian and North American cultivars in integrated management systems for leaf spot of peanut (Arachis hypogaea)[J].Crop Protection, 201 30(6): 698-704.
[27]Kelly L A, Ryley M J, Trevorrow P R, et al. Reduced fungicide use on a new Australian peanut cultivar, highly resistant to the late leaf spot and rust pathogens[J].Australasian Plant Pathology, 2012, 41(4): 359-373.
[28]Pasupuleti J, Ramaiah V, Rathore A, et al. Genetic analysis of resistance to late leaf spot in interspecific groundnuts[J].Euphytica, 2013, 193(1): 13-25.
[29]黎穗临. 广东省花生品种资源研究概述[J].花生科技, 1999(S1): 179-184.
[30]董炜博, 石延茂, 赵志强,等. 花生品种(系)叶部病害综合抗性鉴定[J].中国油料作物学报, 2000(3): 72-75.
[31]袁虹霞, 孙炳剑, 李洪连, 等. 花生品种(系)对叶斑病的抗性鉴定[J].河南農业科学, 2004(12): 35-38.
[32]Sharief Y, Rawlings J O, Gregory W C. Estimates of leaf spot resistance in three interspecific hybrids of Arachis[J].Euphytica, 1978, 27(3): 741-751.
[33]Nevill D J. Inheritance of resistance to Cercosporidium personatum in groundnuts: a genetic model and its implications for selection[J].Oleagineux, 1982, 37(7): 355-363.
[34]Soriano J D, Villanueva A C, Calaguas A M, et al. Gene expression in peanut mutants resistant to leaf spot disease[J].Cell Biology International Reports, 1990, 14: 180.
[35]夏友霖, 敬昱霖, 毛金雄, 等. 花生晚斑病抗性遺传分离分析[J].中国油料作物学报, 2015, 37(2): 134-140.
[36]夏友霖, 廖伯寿, 李加纳, 等. 花生晚斑病抗性AFLP标记[J].中国油料作物学报, 2007(3): 318-321.
[37]Leal-Bertioli S C M, Jose A C V F, Alves-Freitas D M T, et al. Identification of candidate genome regions controlling disease resistance in Arachis[J].BMC Plant Biology, 2009, 9:112.
[38]Khedikar Y P, Gowda M V C, Sarvamangala C, et al. A QTL study on late leaf spot and rust revealed one major QTL for molecular breeding for rust resistance in groundnut (Arachis hypogaea L.)[J].Theoretical and Applied Genetics, 2010, 121(5): 971-984.
[39]Sujay V, Gowda M V C, Pandey M K, et al. Quantitative trait locus analysis and construction of consensus genetic map for foliar disease resistance based on two recombinant inbred line populations in cultivated groundnut (Arachis hypogaea L.)[J].Molecular Breeding, 2012, 30(2): 773-788.
[40]Shoba D, Manivannan N, Vindhiyavarman P, et al. SSR markers associated for late leaf spot disease resistance by bulked segregant analysis in groundnut (Arachis hypogaea L.)[J].Euphytica, 2012, 188(2): 265-272.
[41]Wang H, Pandey M K, Qiao L, et al. Genetic mapping and quantitative trait loci analysis for disease resistance using F2 and F5 generation-based genetic maps derived from ‘TifrunnerבGT-C20 in peanut[J].Plant Genome, 2013, 6(3):1-10.
[42]Pandey M K, Wang H, Khera P, et al. Genetic dissection of novel QTLs for resistance to leaf spots and tomato spotted wilt virus in peanut (Arachis hypogaea L.)[J].Frontiers in Plant Science, 2017, 8: 1-12.
[43]Khera P, Pandey M K, Wang H, et al. Mapping quantitative trait loci of resistance to tomato spotted wilt virus and leaf spots in a recombinant inbred line population of peanut (Arachis hypogaea L.) from SunOleic 97R and NC94022[J].PLoS ONE, 2016, 11(7): 1-17.
[44]Zhou X, Xia Y, Ren X, et al. Construction of a SNP-based genetic linkage map in cultivated peanut based on large scale marker development using next generation double-digest-restriction-site-associated DNA sequencing (ddRADseq)[J].BMC Genomics, 2014, 15: 351.
[45]Zhou X, Xia Y, Liao J, et al. Quantitative trait locus analysis of late leaf spot resistance and plant-type-related traits in cultivated peanut (Arachis hypogaea L.) under multi-environments[J].PLoS ONE, 2016, 11(11): e0166873.
[46]Pandey M K, Khan A W, Singh V K, et al. QTL-seq approach identified genomic regions and diagnostic markers for rust and late leaf spot resistance in groundnut (Arachis hypogaea L.)[J].Plant Biotechnology Journal, 2017, 15(8): 927-941.
[47]Clevenger J, Chu Y, Chavarro C, et al. Mapping late leaf spot resistance in peanut (Arachis hypogaea) using QTL-seq reveals markers for marker-assisted selection[J].Frontiers in Plant Science, 2018, 9: 83.
[48]Chu Y, Chee P, Culbreath A, et al. Major QTLs for resistance to early and late leaf spot diseases are identified on chromosomes 3 and 5 in peanut (Arachis hypogaea)[J].Frontiers in Plant Science, 2019, 10: 883.
[49]Liang Y, Baring M, Wang S, et al. Mapping QTLs for leaf spot resistance in peanut using SNP-based next-generation sequencing markers[J].Plant Breeding and Biotechnology, 2017, 5(2): 115-122.
[50]Lu Q, Liu H, Hong Y, et al. Consensus map integration and QTL meta-analysis narrowed a locus for yield traits to 0.7 cM and refined a region for late leaf spot resistance traits to 0.38 cM on linkage group A05 in peanut (Arachis hypogaea L.)[J].BMC Genomics, 2018, 19(1): 887-897.
[51]Han S, Yuan M, Clevenger J P, et al. A SNP-based linkage map revealed QTLs for resistance to early and late leaf spot diseases in peanut (Arachis hypogaea L.)[J].Frontiers in Plant Science, 2018, 9: 1-9.
[52]Agarwal G, Clevenger J, Pandey M K, et al. High-density genetic map using whole-genome resequencing for fine mapping and candidate gene discovery for disease resistance in peanut[J].Plant Biotechnology Journal, 2018, 16(11): 1954-1967.
[53]Luo M, Dang P, Bausher M G, et al. Identification of transcripts involved in resistance responses to leaf spot disease caused by Cercosporidium personatum in peanut (Arachis hypogaea)[J].Phytopathology, 2005, 95(4): 381-387.
[54]Kumar K R R, Kirti P B. Differential gene expression in Arachis diogoi upon interaction with peanut late leaf spot pathogen, Phaeoisariopsis personata and characterization of a pathogen induced cyclophilin[J].Plant Molecular Biology, 201 75(4/5): 497-513.(下轉第188页)
[55]Guimar[KG-*3]a[DD(-*2]~[][DD)][KG-*6]es P M, Brasileiro A C, Morgante C V, et al. Global transcriptome analysis of two wild relatives of peanut under drought and fungi infection[J].BMC Genomics, 2012, 13(1): 1-15.
[56]Vasavirama K, Kirti P B. Increased resistance to late leaf spot disease in transgenic peanut using a combination of PR genes[J].Functional & Integrative Genomics, 2012, 12(4): 625-634.
[57]Anuradha T S, Divya K, Jami S K, et al. Transgenic tobacco and peanut plants expressing a mustard defensin show resistance to fungal pathogens[J].Plant Cell Reports, 2008, 27(11): 1777-1786.
[58]Iqbal M M, Nazir F, Ali S, et al. Overexpression of rice chitinase gene in transgenic peanut (Arachis hypogaea L.) improves resistance against leaf spot[J].Molecular Biotechnology, 2012, 50(2): 129-136.
[59]Rohini V K, Rao K S. Transformation of peanut (Arachis hypogaea L.) with tobacco chitinase gene: variable response of transformants to leaf spot disease[J].Plant Science, 200 160(5): 889-898.
[60]Tallury S P, Isleib T G, Stalker H T. Comparison of virginia-type peanut cultivars and interspecific hybrid derived breeding lines for leaf spot resistance, yield, and grade[J].Peanut Science, 2009, 36(2): 144-149.
[61]Holbrook C C, Culbreath A K. Registration of ‘Tifrunner peanut[J].J. Plant Regist., 2007, 1(2): 24-58.
[62]Holbrook C C, Stalker H T. Peanut breeding and genetic resources[J].Plant Breeding Reviews, 2010, 22: 297-356.
[63]Holbrook C C, Culbreath A K. Registration of ‘Georganic peanut[J].J. Plant Regist., 2008, 2(1): 17.
收稿日期:2019-08-11
基金項目:国家自然科学基金-国际合作项目(3181101550);山东省重点研发计划项目(2018GNC110036);山东省农业科学院农业科技创新工程项目(CXGC2016B02);河南省科技攻关计划项目(182102110137)
作者简介:吴丽军(1987—),女,助理研究员,主要从事分子生物学研究。E-mail: wljd126@126.com
通讯作者: 袁美(1972—),女,博士,研究员,主要从事花生生物技术育种研究。E-mail: yuanbeauty@126.com
王兴军(1966—),男,博士,研究员,主要从事作物分子育种研究。E-mail: xingjunw@hotmail.com
