CRISPR/Cas基因编辑系统在水稻中的研究进展
2023-07-10刘建菊肖宁吴云雨蔡跃潘存红时薇陈梓春朱书豪李育红余玲王志平刘广青周长海黄年生张小祥季红娟李爱宏
刘建菊 肖宁 吴云雨 蔡跃 潘存红 时薇 陈梓春 朱书豪 李育红 余玲 王志平 刘广青 周长海 黄年生 张小祥 季红娟 李爱宏

摘要:基因编辑是一种能对特定基因进行修饰的基因工程技术,能快速对靶点基因编辑,是高效捕获目的基因、快速研究目标基因功能的重要手段,在基因功能研究和作物育种等方面有着重要意义和广阔的应用前景。基因编辑利用特异的DNA结合元件和切割元件开展编辑工作,然而该技术最需注意的是特异性和脱靶率问题,不同时期的基因编辑技术也针对上述2个问题进行改良,目前应用最为广泛的是CRISPR/Cas9,Cas12a 由于其特异性高且脱靶率大大降低也受到越来越多的关注。本文对基因编辑的技术发展及特点、CRISPR/Cas9和Cas12a的技术优势进行介绍,并对这2种技术在水稻产量、抗性及品质中的研究进展进行综述,同时对拓展CRISPR/Cas基因编辑技术在水稻中的应用提出展望,为基因功能鉴定及遗传改良提供参考。
关键词:基因编辑;Cas9;Cas12a;水稻;性状改良
中图分类号:S511.01文献标志码:A文章编号:1002-1302(2023)11-0001-09
基因编辑(gene editing)是一种能对特定基因进行修饰的基因工程技术[1-2],该技术利用工程核酸酶切割目标基因组产生DNA双链断裂(DSB),进而激活细胞内源性DNA修复机制从而产生包括插入、缺失及基因片段替换等新的基因突变类型[3-5]。
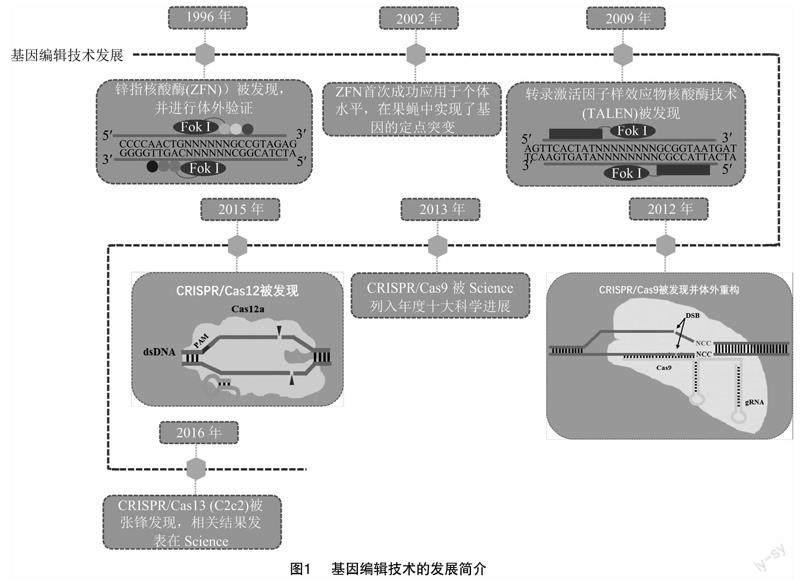
1996年出现的锌指核酸酶(ZFN)为基因编辑技术的发展奠定了基础[6-7],利用该技术首次于2002年果蝇染色体上实现基因定点突变[8]。随后转录激活样效应因子核酸酶(TALENs)[9]及由RNA介导的Cas9蛋白相关的成簇规则间隔短回文重复序列(CRISPR)相繼被发现[10-11],特别是CRISPR/Cas9于2013年开始应用于植物基因组编辑,被Science列入2013年十大科学进展[10]。此外,用于切割双链DNA的CRISPR/Cas12a(Cpf1)[12-13]及在crRNA指导下切割ssRNA的CRISPR/Cas13(C2c2)[14]于2015年和2016年相继被发现(图1)。
基因编辑利用特异的DNA结合元件和切割元件开展编辑工作,然而该技术最需注意的是特异性和脱靶率问题,基因编辑技术的更迭对这2个方面的改善也各不相同(表1)。ZFNs是第一个应用于基因定点编辑的技术,然而其ZFN 剪切DNA 形成同源二聚体的同时,可能会产生异源二聚体引起脱靶且难以实现多靶点编辑等问题,严重阻碍了其应用[15-16];TALENs技术是1个TALE基序识别1个碱基对,因此多个串联的TALE基序与其识别的碱基对呈一一对应关系,大大提高了编辑特异性并降低脱靶率,但其编辑效率较低,且难以进行多基因编辑[17-20];CRISPR/Cas9技术在sgRNA的指导下与靶点结合,并利用HNH和RuvC对外源DNA进行切割,其编辑效率大大提高,且可以对多基因同时编辑,然而其缺点是靶向目标 DNA 序列容易出现错配,存在脱靶率高、编辑特异性低等缺陷[4,16,21-22];Cas12a可以在crRNA引导下识别PAM,识别到正确序列才会形成封闭的R环,因此编辑准确性相对Cas9有了较大提高,其脱靶率也有所降低[12-13,23]。
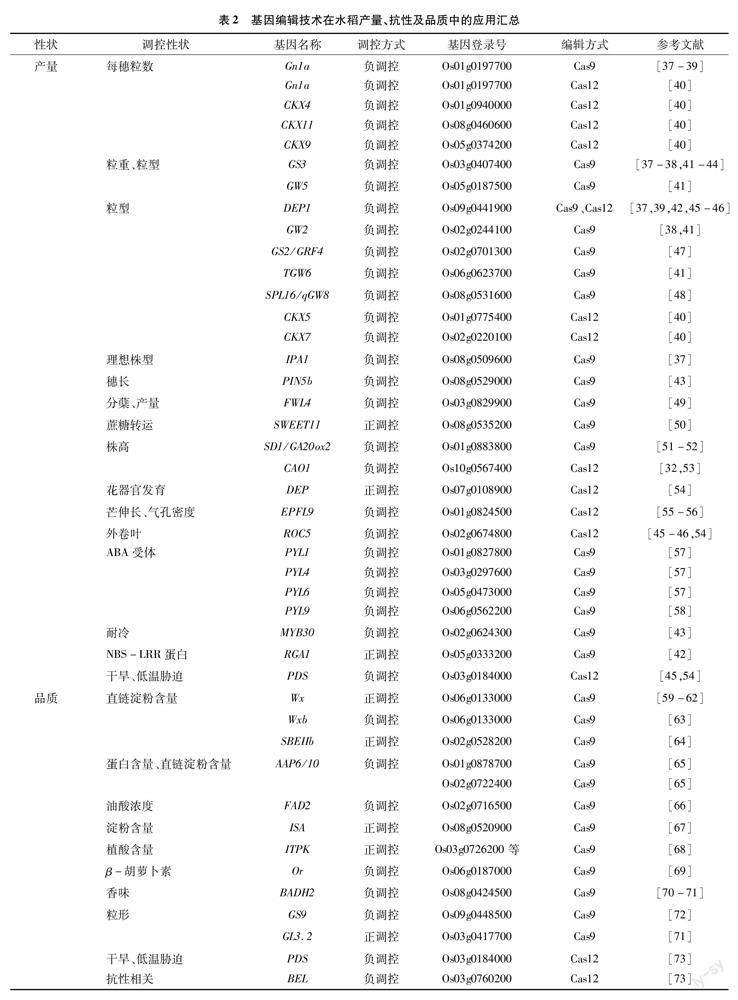
CRISPR/Cas9及Cas12a是目前基因编辑技术中应用最为广泛的2种技术,在水稻产量、品质、生物胁迫及非生物胁迫性状关键基因的分子遗传功能解析和目标性状的精准改良上已成熟应用(表2)。
2CRISPR/Cas在水稻中的研究进展
2.1产量性状
水稻产量由单株穗数、每穗粒数、粒型及粒重等多个性状综合组成[112-113]。目前已有29个产量相关基因被编辑,其中4个基因对产量起正调控作用,其他25个基因均作为负调控因子发挥作用。Li等对每穗粒数Gn1a、粒型DEP1、粒重GS3及理想株型基因IPA1定点突变,gn1a、dep1和gs3的T2突变体出现穗粒数增加、粒型变大,成功提高了产量[37]。其他研究分别对Gn1a&DEP1、GS3&DEP1、GS3、GS2/GRF4及SPL16/qGW8等开展基因编辑,在穗粒数、粒型、粒重等性状上调控产量,改善农艺性状同时提高产量[39,42,44,47-48]。开展多基因同时编辑也可快速调控产量,Xu等同时对负调控粒重、粒型基因GS3、GW2、GW5及TGW6进行编辑,快速改良突变体粒重及产量[41]。Zhou等同时编辑GS3、Gn1a及GW2,相关突变体出现籽粒变大、穗粒数增多从而提高水稻产量[38]。Zeng等同时编辑PIN5b、GS3和MYB30,突变体兼顾了高产和耐冷性[43]。非产量调控基因突变也会提高产量,Miao等获得ABA受体突变体pyl1/4/6,通过增加31%籽粒数量从而提高产量[57],除此之外,对FWL4、SD1(OsGA20ox2)及PYL9进行定点突变也可不同程度提高产量[49,51-52,58]。然而产量正调控基因如RGA1、SWEET11被编辑后会分别引起植株极端矮化及灌浆功能受损,从而减产[42,50]。
CRISPR/Cas12a在水稻产量调控中应用也日渐增多,Malzahn等对粒长基因DEP1和叶片卷曲度基因ROC5进行敲除提高产量。对水稻PDS、DEP 和ROC5基因所有靶点进行突变,能同时改良农艺性状及抗性[45,54],而将叶绿素a加氧酶基因CAO1靶向敲入水稻中,突变体的产量及品质降低[32,53],Zheng等同时利用Cas9和Cas12a对细胞分裂素家族基因OsCKX1-11进行编辑,获得了农艺性状及产量均有提升的单基因及多基因突变体,Cas9的编辑效率为26.9%~90.0%,有8个基因的编辑效率高于50.0%,而Cas12a的编辑效率为368%~100%且9个基因的编辑效率高于60%,Cas12a的多基因编辑效率高于Cas9(91.7%>545%)[40]。上述研究表明,对负调控基因进行定点突变后可快速获得目标性状改善的编辑系,然而有些基因突变后会对其他性状产生不利影响,因此多重基因编辑技术的应用为多个性状同时改良提供了方案和可行性,在开展基因编辑时Cas12a的编辑效率及稳定性均高于Cas9。
2.2品质性状
稻米品质是水稻商业价值的核心卖点,受到多个基因综合调控,已有大量基因被证实直接或间接调控稻米品质,可用于定向改良直链淀粉含量、蛋白、香味等性状。目前有13个品质基因被编辑,其中4个基因(ISA、ITPK、GL3.2和BEL)正调控稻米品质,其他基因负调控稻米品质。Wx基因的基因编辑位置差异对稻米品质影响不同,对Wx基因功能位点进行突变,可以将直链淀粉含量降至与糯稻相似,在不影响产量前提下改良稻米品质[59-61];对 Wxb基因启动子转录因子结合位点进行突变,获得新的Wx等位基因并获得直链淀粉含量不同程度降低的突变体,改良了稻米品质[62]。fad2突变体的油酸浓度提高,gs9突变体的粒型、垩白及外观等品质显著改善,or突变体籽粒β-胡萝卜素含量显著提高,isa突变体总淀粉含量下调,ZmPsy和SSU-crtI突变体水稻的籽粒类胡萝卜素含量提高,badh2突变体籽粒产生香味,均可改良稻米品质[66-67,69-70,72,114]。多基因同時突变可综合提升水稻性状,如app6/10双突变体的直链淀粉、蛋白及谷蛋白含量均下调[65];细胞色素P450家族基因(Os03g0603100、Os03g0568400和GL3.2)和香味基因BADH2同时突变后改良稻米香味并提高产量[71];PDS和BELs同时突变稳定提高水稻产量和品质[73]。对正调控基因进行突变,有助于理解基因在稻米品质改良中的作用,敲除Wxb第一内含子、SBEIIb进行精准敲除,突变体直链淀粉含量上调,且引起营养特性改变[63-64]。Jiang等突变ITPK1-6,降低籽粒植酸含量然而却提高无机磷含量,不利于水稻生长繁殖,证实该基因对水稻正常生长发育的重要性[68]。对负调控稻米品质基因的敲除加速了优质水稻品种选育的进程,与其他产量性状相关基因同时编辑,有望在保证产量的同时提高品质。
2.3生物胁迫
水稻生长过程对生物胁迫的抗性也可利用基因编辑方法改良,对抗性相关基因MPK1、MPK2、MPK5和MPK6的敲除能够提高抗病性[85-86]。ERF922、SEC3A、ALB1、RSY1 和Pi21敲除后,突变体对稻瘟病的抗性提高,同时农艺性状也得到改良[74-78]。SWEET13和SWEET14敲除后突变体对白叶枯病菌的抗性提高,且SWEET14突变体无产量损失[79,81]。对SWEET11/8N3/Xa13编码区及启动子区定点突变,也能提高水稻对白叶枯病的抗性[80,82]。Liang等对稻曲病相关基因USTA和UvSLT2进行编辑,显著提高了水稻对稻曲病抗性[84]。利用Cas12a低水平同源性核酸酶MAD7对水稻基因EPSPS、NRAMP、PDS、Xa13及ALS等进行多重基因敲除,同步提升了突变体的品质、除草剂及白叶枯病抗性[83]。Wang等利用Cas12a对受体样激酶(OsRLK)相关基因(OsRLK-798、OsRLK-799、OsRLK-802和OsRLK-803)及CYP81A家族基因(OsBEL-230、OsBEL-240、OsBEL-250和OsBEL-260)开展多重基因编辑,获得了阳性植株,相关突变体调控了水稻的抗逆性[105]。
对水稻负调控抗性基因进行敲除或替换可快速改善目标性状,提升水稻抗性,然而有些编辑以损失产量为代价[109],而有些编辑在不损害甚至优化农艺性状前提下同步改善水稻品质[77-78,81,90,95],因此在进行水稻抗性改良时需要考虑基因对水稻的综合影响,从而制定相应编辑策略。
2.4非生物胁迫
水稻生长发育过程中会受到多种非生物胁迫的影响,如干旱、低温、盐、除草剂等,相关基因的大量挖掘促进了基因编辑在水稻非生物胁迫中的应用,目前有24个相关基因被编辑,其中8个基因起正调控作用,即Ann3、OTS1、RAV2、SAPK2、BELs、MKK5、RLKs和SAP。在水稻抗旱性方面,PYL9、ERA1、PDS、半卷叶基因(SRL1和SRL2)和MIR535的基因突变会增强突变体的抗旱性[58,88-90,106]。而敲除SAPK2和SAP基因后,突变体对干旱胁迫和活性氧更敏感,农艺性状显著下降[87,111]。在水稻响应盐胁迫方面,敲除水稻中的RR22、DST及PQT3基因,可显著提高耐盐性且不影响农艺性状[92,94-95],但对OTS1编码区及RAV2启动子的GT-1元件突变后,其耐盐性下降[91,93]。在水稻抗除草剂方面,通过将EPSPS、ALS突变基因敲入,或点突变野生型基因(ALS、FTIP1e)均能使水稻获得除草剂抗性[96-103]。
除此之外,敲除Nramp5能降低Cd的积累且不影响产量[107-108];Ann3敲除后对低温的耐受性降低[110];敲除MKK5后,突变体抗逆性降低[104];同时突变抽穗基因Hd2、Hd4和Hd5后突变体开花期及成熟期提前有助于逃避胁迫[109],然而农艺性状受到较大影响,因此在应用时可进行单基因编辑,从而消除对产量的损害。
3CRISPR/Cas的技术展望
基因编辑技术为生命科学带来重大进展,然而几种技术的脱靶率及特异性问题仍需重点关注。研究人员优化了相关技术,开发了DB-PACE法从而降低基因编辑工具酶的脱靶效应,大大提高TALEN核酸酶的DNA结合能力和切割特异性[115];开发出提高Cas9基因编辑和碱基编辑特异性的选择性核输出抑制剂(SINE)[116];Sheng利用腙介导CRISPR/Cas12a系统,通过互补碱基配对引起的邻近效应来加速整个激活链的形成,从而提高Cas12a 系统的特异性[117]。除此之外,CRISPR系统的sgRNA的优化、PAM修饰、crRNA优化及Cas蛋白突变体挖掘也会进一步提高编辑范围及特异性并降低脱靶率[12,46,104,118-120]。此外Cas12a蛋白表现出对低温敏感的特征,目前Cas12a突变体是解决该问题的主要方式,而引起低温敏感的分子机制尚不明确。上述问题的解决,将大大提高基因编辑水平,对目标基因进行定向编辑,产生无外源DNA插入的新品种,从而加快育种速度、缩短育种年限。
水稻產量、抗性和品质相关基因的挖掘及分子机理解析,有助于更全面了解基因功能,目前基因编辑主要集中在编码区,有少量研究是编辑启动子的转录结合位点实现性状调控的。已有研究表明,DNA结构本身,如拓扑异构结构等也会影响基因表达水平[121],因此,未来也可能作为基因编辑靶点,增加目标性状精准改良的可能性。随着人工智能的发展,Alphafold等技术对蛋白预测精准度提高,越来越多的蛋白结构被预测,对目标基因的模拟突变有助于挖掘关键碱基序列,可进行靶向预测,实现新的目标性状的改良已经成为可能。相信随着基因编辑技术的不断完善、生物信息学和人工智能的不断发展,水稻育种将会迅猛发展。
参考文献:
[1]Yin K,Gao C,Qiu J L. Progress and prospects in plant genome editing[J]. Nature Plants,2017,3(8):1-6.
[2]李君,张毅,陈坤玲,等. CRISPR/Cas 系统:RNA 靶向的基因组定向编辑新技术[J]. 遗传,2013,35(11):1265-1273.
[3]Kim H,Kim J S. A guide to genome engineering with programmable nucleases[J]. Nature Reviews Genetics,2014,15(5):321-334.
[4]Shan Q,Wang Y,Li J,et al. Targeted genome modification of crop plants using a CRISPR-Cas system[J]. Nature Biotechnology,2013,31(8):686-688.
[5]张白雪,孙其信,李海峰. 基因修饰技术研究进展[J]. 生物工程学报,2015,31(8):1162-1174.
[6]Urnov F D,Miller J C,Lee Y L,et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases[J]. Nature,2005,435(7042):646-651.
[7]Miller J C,Holmes M C,Wang J,et al. An improved zinc-finger nuclease architecture for highly specific genome editing[J]. Nature Biotechnology,2007,25(7):778-785.
[8]Bibikova M,Golic M,Golic K G,et al. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases[J]. Genetics,2002,161(3):1169-1175.
[9]Boch J,Scholze H,Schornack S,et al. Breaking the code of DNA binding specificity of TAL-type Ⅲ effectors[J]. Science,2009,326(5959):1509-1512.
[10]Jinek M,Chylinski K,Fonfara I,et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity [J]. Science,2012,337(6096):816-821.
[11]Gasiunas G,Barrangou R,Horvath P,et al. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria[J]. Proceedings of the National Academy of Sciences,2012,109(39):E2579-E2586.
[12]Makarova K S,Koonin E V. Annotation and classification of CRISPR-Cas systems[J]. CRISPR:Methods and Protocols,2015,1311:47-75.
[13]Zetsche B,Gootenberg J S,Abudayyeh O O,et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system[J]. Cell,2015,163(3):759-771.
[14]Abudayyeh O O,Gootenberg J S,Konermann S,et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector[J]. Science,2016,353(6299):aaf5573.
[15]Ramirez C L,Foley J E,Wright D A,et al. Unexpected failure rates for modular assembly of engineered zinc fingers[J]. Nature Methods,2008,5(5):374-375.
[16]Gupta R M,Musunuru K. Expanding the genetic editing tool kit:ZFNs,TALENs,and CRISPR-Cas9 [J]. The Journal of Clinical Investigation,2014,124(10):4154-4161.
[17]Reyon D,Tsai S Q,Khayter C,et al. FLASH assembly of TALENs for high-throughput genome editing[J]. Nature Biotechnology,2012,30(5):460-465.
[18]Kim Y,Kweon J,Kim A,et al. A library of TAL effector nucleases spanning the human genome [J]. Nature Biotechnology,2013,31(3):251-258.
[19]Guilinger J P,Pattanayak V,Reyon D,et al. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity[J]. Nature Methods,2014,11(4):429-435.
[20]Smith C,Gore A,Yan W,et al. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs[J]. Cell Stem Cell,2014,15(1):12-13.
[21]Cong L,Ran F A,Cox D,et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science,2013,339(6121):819-823.
[22]Nekrasov V,Staskawicz B,Weigel D,et al. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease[J]. Nature Biotechnology,2013,31(8):691-693.
[23]Zeng Y,Hong Y,Azi F,et al. Advanced genome-editing technologies enable rapid and large-scale generation of genetic variants for strain engineering and synthetic biology[J]. Current Opinion in Microbiology,2022,69:102175.
[24]Yang G,Huang X. Methods and applications of CRISPR/Cas system for genome editing in stem cells[J]. Cell Regeneration,2019,8(2):33-41.
[25]Osakabe K,Osakabe Y,Toki S. Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases[J]. Proceedings of the National Academy of Sciences,2010,107:12034-12039.
[26]Miller J C,Tan S,Qiao G,et al. A TALE nuclease architecture for efficient genome editing[J]. Nature Biotechnology,2011,29:143-150.
[27]趙钦军,韩忠朝. 基因编辑技术的发展前景及伦理与监管问题探讨[J]. 科学与社会,2016,6(3):1-11.
[28]Sood R,Carrington B,Bishop K,et al. Efficient methods for targeted mutagenesis in zebrafish using zinc-finger nucleases:data from targeting of nine genes using CompoZr or CoDA ZFNs[J]. PloS One,2013,8(2):e57239.
[29]Arazoe T,Ogawa T,Miyoshi K,et al. Tailor‐made TALEN system for highly efficient targeted gene replacement in the rice blast fungus[J]. Biotechnology and Bioengineering,2015,112(7):1335-1342.
[30]Naeem M,Majeed S,Hoque M Z,et al. Latest developed strategies to minimize the off-target effects in CRISPR-Cas-mediated genome editing[J]. Cells,2020,9(7):1608.
[31]Hruscha A,Krawitz P,Rechenberg A,et al. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish [J]. Development,2013,140(24):4982-4987.
[32]Endo A,Masafumi M,Kaya H,et al. Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida[J]. Scientific Reports,2016,6(1):38169.
[33]Miller J C,Patil D P,Xia D F,et al. Enhancing gene editing specificity by attenuating DNA cleavage kinetics[J]. Nature Biotechnology,2019,37(8):945-952.
[34]Wang X,Wang Y,Wu X,et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors[J]. Nature Biotechnology,2015,33(2):175-178.
[35]Khandagale K,Nadaf A. Genome editing for targeted improvement of plants[J]. Plant Biotechnology Reports,2016,10:327-343.
[36]Kim H K,Song M,Lee J,et al. In vivo high-throughput profiling of CRISPR-Cpf1 activity [J]. Nature Methods,2017,14(2):153-159.
[37]Li M,Li X,Zhou Z,et al. Reassessment of the four yield-related genes Gn1a,DEP1,GS3,and IPA1 in rice using a CRISPR/Cas9 system [J]. Frontiers in Plant Science,2016,7:377.
[38]Zhou J,Xin X,He Y,et al. Multiplex QTL editing of grain-related genes improves yield in elite rice varieties[J]. Plant Cell Reports,2019,38:475-485.
[39]Huang L,Zhang R,Huang G,et al. Developing superior alleles of yield genes in rice by artificial mutagenesis using the CRISPR/Cas9 system [J]. The Crop Journal,2018,6:475-481.
[40]Zheng X,Zhang S,Liang Y,et al. Loss-function mutants of OsCKX gene family based on CRISPR-Cas systems revealed their diversified roles in rice[J]. The Plant Genome,2023,e20283.
[41]Xu R,Yang Y,Qin R,et al. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice[J]. Journal of Genetics and Genomics,2016,43(8):529-532.
[42]Cui Y,Jiang N,Xu Z,et al. Heterotrimeric G protein are involved in the regulation of multiple agronomic traits and stress tolerance in rice[J]. BMC Plant Biology,2020,20:1-13.
[43]Zeng Y,Wen J,Zhao W,et al. Rational improvement of rice yield and cold tolerance by editing the three genes OsPIN5b,GS3,and OsMYB30 with the CRISPR-Cas9 system[J]. Front Plant Science,2020,10:1663.
[44]Huang J,Gao L,Luo S,et al. The genetic editing of GS3 via CRISPR/Cas9 accelerates the breeding of three-line hybrid rice with superior yield and grain quality [J]. Molecular Breeding,2022,42(4):22.
[45]Tang X,Lowder,Zhang T,et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants[J]. Nature Plants,2017,3:17018.
[46]Malzahn A A,Tang X,Lee K,et al. Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice,maize,and Arabidopsis[J]. BMC Biology,2019,17(1):1-14.
[47]Wang W,Wang W,Pan Y,et al. A new gain-of-function OsGS2/GRF4 allele generated by CRISPR/Cas9 genome editing increases rice grain size and yield[J]. The Crop Journal,2022,10(4):1207-1212.
[48]Usman B,Nawaz G,Zhao N,et al. Programmed editing of rice (Oryza sativa L.) OsSPL16 gene using CRISPR/Cas9 improves grain yield by modulating the expression of pyruvate enzymes and cell cycle proteins[J]. International Journal of Molecular Sciences,2020,22(1):249.
[49]Gao Q,Li G,Sun H,et al. Targeted mutagenesis of the rice FW 2.2-like gene family using the CRISPR/Cas9 system reveals OsFWL4 as a regulator of tiller number and plant yield in rice[J]. International Journal of Molecular Sciences,2020,21(3):809.
[50]Ma L,Zhang D,Miao Q,et al. Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling[J]. Plant and Cell Physiology,2017,58(5):863-873.
[51]Hu X,Cui Y,Dong G,et al. Using CRISPR-Cas9 to generate semi-dwarf rice lines in elite landraces [J]. Scientific Reports,2019,9:19096.
[52]Han Y,Teng K,Nawaz G,et al. Generation of semi-dwarf rice (Oryza sativa L.) lines by CRISPR/Cas9-directed mutagenesis of OsGA20ox2 and proteomic analysis of unveiled changes caused by mutations[J]. 3 Biotech,2019,9:387.
[53]Begemann M B,Gray B N,January E,et al. Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases[J]. Scientific Reports,2017,7(1):11606.
[54]Mahfouz M M. Genome editing:the efficient tool CRISPR-Cpf1[J]. Nature Plants,2017,3(3):1-2.
[55]Yin X,Biswal A K,Dionora J,et al. CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice[J]. Plant Cell Reports,2017,36:745-757.
[56]Yin X,Anand A,Quick P,et al. Editing a stomatal developmental gene in rice with CRISPR/Cpf1[J]. Plant Genome Editing with CRISPR Systems:Methods and Protocols,2019,257-268.
[57]Miao C,Xiao L,Hua K,et al. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity[J]. Proceedings of the National Academy of Sciences,2018,115:6058-6063.
[58]Usman B,Nawaz G,Zhao N,et al. Precise editing of the OsPYL9 gene by RNA-guided Cas9 nuclease confers enhanced drought tolerance and grain yield in rice (Oryza sativa L.) by regulating circadian rhythm and abiotic stress responsive proteins[J]. International Journal of Molecular Sciences,2020,21:7854.
[59]Ma X,Zhang Q,Zhu Q,et al. A robust CRISPR/Cas9 system for convenient,high-efficiency multiplex genome editing in monocot and dicot plants[J]. Molecular Plant,2015,8(8):1274-1284.
[60]Zhang J,Zhang H,Botella J R,et al. Generation of new glutinous rice by CRISPR/Cas9‐targeted mutagenesis of the Waxy gene in elite rice varieties[J]. Journal of Integrative Plant Biology,2018,60(5):369-375.
[61]Fei Y Y,Jie Y,Wang F Q,et al. Production of two elite glutinous rice varieties by editing wx gene[J]. Rice Science,2019,26(2):118-124.
[62]Huang L,Li Q,Zhang C,et al. Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system [J]. Plant Biotechnology Journal,2020,18:2164 -2166.
[63]Liu X,Ding Q,Wang W,et al. Targeted deletion of the first intron of the Wxb allele via CRISPR/Cas9 significantly increases grain amylose content in rice[J]. Rice,2022,15:1-12.
[64]Sun Y,Jiao G,Liu Z,et al. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes[J]. Frontiers in Plant Science,2017,8:298.
[65]Wang S,Yang Y,Guo M,et al. Targeted mutagenesis of amino acid transporter genes for rice quality improvement using the CRISPR/Cas9 system[J]. The Crop Journal,2020,8:457-464.
[66]Abe K,Araki E,Suzuki Y,et al. Production of high oleic/low linoleic rice by genome editing[J]. Plant Physiology and Biochemistry,2018,131:58-62.
[67]Chao S F,Cai Y C,Feng B B,et al. Editing of rice isoamylase gene ISA1 provides insights into its function in starch formation[J]. Rice Science,2019,26:77-87.
[68]Jiang M,Liu Y,Liu Y,et al. Mutation of inositol 1,3,4-trisphosphate 5/6-kinase6 impairs plant growth and phytic acid synthesis in rice [J]. Plants,2019,8(5):114.
[69]Endo A,Saika H,Takemura M,et al. A novel approach to carotenoid accumulation in rice callus by mimicking the cauliflower Orange mutation via genome editing[J]. Rice,2019,12(1):1-5.
[70]Ashokkumar S,Jaganathan D,Ramanathan V,et al. Creation of novel alleles of fragrance gene OsBADH2 in rice through CRISPR/Cas9 mediated gene editing[J]. PloS One,2020,15(8):e0237018.
[71]Usman B,Nawaz G,Zhao N,et al. Generation of high yielding and fragrant rice (Oryza sativa L.) lines by CRISPR/Cas9 targeted mutagenesis of three homoeologs of Cytochrome P450 gene family and OsBADH2 and transcriptome and proteome profiling of revealed changes triggered by mutations[J]. Plants,2020,9:788.
[72]Zhao D S,Li Q F,Zhang C Q,et al. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality[J]. Nature Communication,2018,9:1240.
[73]Xu R F,Qin R Y,Li H,et al. Generation of targeted mutant rice using a CRISPR-Cpf1 system[J]. Plant Biotechnology Journal,2017,15(6):713-717.
[74]Wang F,Wang C,Liu P,et al. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922[J]. PloS One,2016,11:1-18.
[75]Ma J,Chen J,Wang M,et al. Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice[J]. Journal of Experimental Botany,2017,69:1051-1064.
[76]Foster A J,Martin-Urdiroz M,Yan X,et al. CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus[J]. Scientific Reports,2018,8:14355.
[77]Li S,Shen L,Hu P,et al. Developing disease-resistant thermosensitive male sterile rice by multiplex gene editing [J]. Journal of Integrative Plant Biology,2019,61:1201-1205.
[78]Nawaz G,Usman B,Peng H,et al. Knockout of Pi21 by CRISPR/Cas9 and iTRAQ-based proteomic analysis of mutants revealed new insights into M. oryzae resistance in elite rice line[J]. Genes,2020,11(7):735.
[79]Zhou J,Peng Z,Long J,et al. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice[J]. The Plant Journal,2015,82(4):632-643.
[80]Kim Y A,Moon H,Park C J. CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae[J]. Rice,2019,12:67.
[81]Zeng X,Luo Y,Vu N T Q,et al. CRISPR/Cas9-mediated mutation of OsSWEET14 in rice cv. Zhonghua11 confers resistance to Xanthomonas oryzae pv. oryzae without yield penalty[J]. BMC Plant Biology,2020,20(1):1-11.
[82]Li C,Li W,Zhou Z,et al. A new rice breeding method:CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice [J]. Plant Biotechnology Journal,2020,18:313-315.
[83]Lin Q,Zhu Z,Liu G,et al. Genome editing in plants with MAD7 nuclease[J]. Journal of Genetics and Genomics,2021,48(6):444-451.
[84]Liang Y,Han Y,Wang C,et al. Targeted deletion of the USTA and UvSLT2 genes efficiently in Ustilaginoidea virens with the CRISPR-Cas9 system [J]. Frontiers in Plant Science,2018,9:699.
[85]Xie K,Yang Y. RNA-guided genome editing in plants using a CRISPR-Cas system[J]. Molecular Plant,2013,6:1975-1983.
[86]Minkenberg B,Xie K,Yang Y. Discovery of rice essential genes by characterizing a CRISPR-edited mutation of closely related rice MAP kinase genes[J]. The Plant Journal,2017,89:636-648
[87]Lou D,Wang H,Liang G,et al. OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice[J]. Frontiers in Plant Science,2017,8:993.
[88]Ogata T,Ishizaki T,Fujita M,et al. CRISPR/Cas9-targeted mutagenesis of OsERA1 confers enhanced responses to abscisic acid and drought stress and increased primary root growth under nonstressed conditions in rice[J]. PLoS One,2020,15(12):e0243376.
[89]Banakar R,Schubert M,Collingwood M,et al. Comparison of CRISPR-Cas9/Cas12a ribonucleoprotein complexes for genome editing efficiency in the rice phytoene desaturase (OsPDS) gene[J]. Rice,2020,13(1):1-7.
[90]Liao S,Qin X,Luo L,et al. CRISPR/Cas9-induced mutagenesis of semi-rolled Leaf1,2 confers curled leaf phenotype and drought tolerance by influencing protein expression patterns and ROS scavenging in rice (Oryza sativa L.) [J]. Agronomy,2019,9(11):728.
[91]Duan Y B,Li J,Qin R Y,et al. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis[J]. Plant Molecular Biology,2016,90:49-62.
[92]Zhang A,Liu Y,Wang F,et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene[J]. Molecular Breeding,2019,39:1-10.
[93]Zhang C,Srivastava A K,Sadanandom A. Targeted mutagenesis of the SUMO protease,Overly Tolerant to Salt1 in rice through CRISPR/Cas9-mediated genome editing reveals a major role of this SUMO protease in salt tolerance[J]. BioRxiv,2019:555706.
[94]Santosh Kumar V V,Verma R K,Yadav S K,et al. CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010[J]. Physiology and Molecular Biology of Plants,2020,26:1099-1110.
[95]Alfatih A,Wu J,Jan S U,et al. Loss of rice PARAQUAT TOLERANCE 3 confers enhanced resistance to abiotic stresses and increases grain yield in field[J]. Plant,Cell & Environment,2020,43(11):2743-2754.
[96]Li J,Meng X,Zong Y,et al. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9 [J]. Nature Plants,2016,2(10):1-6.
[97]Shimatani Z,Kashojiya S,Takayama M,et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion[J]. Nature Biotechnology,2017,35:441-443.
[98]Sun Y,Zhang X,Wu C,et al. Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase[J]. Molecular Plant,2016,9(4):628-631.
[99]Kuang Y,Li S,Ren B,et al. Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms [J]. Molecular Plant,2020,13(4):565-572.
[100]Wang F,Xu Y,Li W,et al. Creating a novel herbicide-tolerance OsALS allele using CRISPR/Cas9-mediated gene editing[J]. The Crop Journal,2021,9(2):305-312.
[101]Zhang R,Chen S,Meng X,et al. Generating broad-spectrum tolerance to ALS-inhibiting herbicides in rice by base editing[J]. Science China Life Sciences,2021,64:1624-1633.
[102]Li S,Li J,Zhang J,et al. Synthesis-dependent repair of Cpf1-induced double strand DNA breaks enables targeted gene replacement in rice [J]. Journal of Experimental Botany,2018,69(20):4715-4721.
[103]Li S,Li J,He Y,et al. Precise gene replacement in rice by RNA transcript-templated homologous recombination [J]. Nature Biotechnology,2019,37(4):445-450.
[104]Zhang Q,Yin K,Liu G,et al. Fusing T5 exonuclease with Cas9 and Cas12a increases the frequency and size of deletion at target sites[J]. Science China Life Sciences,2020,63:1918-1927.
[105]Wang M,Mao Y,Lu Y,et al. Multiplex gene editing in rice using the CRISPR-Cpf1 system[J]. Molecular Plant,2017,10(7):1011-1013.
[106]Yue E,Cao H,Liu B. OsmiR535,a potential genetic editing target for drought and salinity stress tolerance in Oryza sativa[J]. Plants,2020,9(10):1337.
[107]Tang L,Mao B,Li Y,et al. Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield[J]. Scientific Reports,2017,7(1):14438.
[108]Yang C H,Zhang Y,Huang C F. Reduction in cadmium accumulation in japonica rice grains by CRISPR/Cas9-mediated editing of OsNRAMP5[J]. Journal of Integrative Agriculture,2019,18(3):688-697.
[109]Li X,Zhou W,Ren Y,et al. High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing [J]. Journal of Genetics and Genomics,2017,44:175-178.
[110]Shen C,Que Z,Xia Y,et al. Knock out of the annexin gene OsAnn3 via CRISPR/Cas9-mediated genome editing decreased cold tolerance in rice[J]. Journal of Plant Biology,2017,60:539-547.
[111]Park J R,Kim E G,Jang Y H,et al. Applications of CRISPR/Cas9 as new strategies for short breeding to drought gene in rice[J]. Frontiers in Plant Science,2022,13.
[112]Wang Y J,Li J Y. Molecular basis of plant architecture[J]. Annual Review of Plant Biology,2008,59:253 -279.
[113]Xing Y,Zhang Q. Genetic and molecular bases of rice yield[J]. Annual Review of Plant Biology,2010,61:421-442.
[114]Dong O X,Yu S,Jain R,et al. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9[J]. Nature Communications,2020,11(1):1178.
[115]Hubbard B P,Badran A H,Zuris J A,et al. Continuous directed evolution of DNA-binding proteins to improve TALEN specificity [J]. Nature Methods,2015,12(10):939-942.
[116]Cui Y R,Wang S J,Ma T,et al. KPT330 improves Cas9 precision genome-and base-editing by selectively regulating mRNA nuclear export[J]. Communications Biology,2022,5(1):237.
[117]Sheng A,Yang J,Tang L,et al. Hydrazone chemistry-mediated CRISPR/Cas12a system for bacterial analysis[J]. Nucleic Acids Research,2022,50(18):10562-10570.
[118]Lee K,Zhang Y,Kleinstiver B P,et al. Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize [J]. Plant Biotechnology Journal,2019,17(2):362-372.
[119]Gao L,Cox D B,Yan W X,et al. Engineered Cpf1 variants with altered PAM specificities[J]. Nature Biotechnology,2017,35(8):789-792.
[120]王敬文,嚴芳,柳浪,等. 水稻 CRISPR/Cas12a 系统的优化及其介导的腺嘌呤碱基编辑器的建立[J]. 生物技术通报,2021,37(6):279.
[121]Oudelaar A M,Higgs D R. The relationship between genome structure and function[J]. Nature Reviews Genetics,2021,22:154-168.
