一种多吡啶双核单功能铂配合物的合成、晶体结构及抗癌活性
2023-02-03李训一王宇静周子默汪清清胡美纯杨晓松王小波
李训一 王宇静 周子默,3 方 霄 汪清清 胡美纯 杨晓松 王小波*,
(1湖北科技学院医学部药学院,咸宁 437100)
(2湖北科技学院医学部,糖尿病心脑血管病变湖北省重点实验室,咸宁 437100)
(3湖北科技学院医学部基础医学院,咸宁 437100)
0 Introduction
Platinum‑based drugs are classic anti‑cancer che‑motherapeutics and are still widely used as first‑line therapy to treat solid neoplasms,mainly because of their broad ‑spectrum anticancer activities,their dis‑tinct therapeutic effects,and their preferable econo‑my[1‑2].Currently,approximately 50% of all chemothera‑py regimens including platinum ‑based drugs are uti‑lized as the main therapy or co‑therapy for clinical can‑cer treatment[3].Commonly used platinum‑based drugs include cisplatin,carboplatin,oxaliplatin,nedaplatin,lobaplatin,and heptaplatin.All of these drugs are square‑planar platinumcomplexes with two leaving groups and two non‑leaving ligands coordinated to the Ptcenter[1].Because of their structural similarity,platinum‑based drugs share many general characteris‑tics.Hence,there are indeed existing some common problems when referring to the advantages and disad‑vantages of platinum‑based drugs.And this clinical dilemma is a serious impediment to the development of new platinum‑based drugs with novel antitumor mecha‑nisms.More unfavorably,platinum‑based drugs gener‑ally demonstrate several serious side effects,including nephrotoxicity,myelosuppression,neurotoxicity,and ototoxicity,which dramatically limit their clinical use[4‑7].Another major obstacle to developing new plati‑num compounds with novel antitumor mechanisms is drug resistance[8‑9].It is therefore of vital importance to optimize the chemical properties,bioactivities and met‑abolic pathways,and mechanism of actions of novel platinum‑based compounds.
Among the strategies used to overcome these adversities,one of the most common approaches is to design and exploit monofunctional platinum complexes which contain only one leaving group(in contrast with traditional platinum drugs)[10‑14].Under this principle,many monofunctional Pt complexes have been thor‑oughly synthesized,and their anticancer activities and mechanisms of action have been investigated[15‑20].In comparison with classical bifunctional drugs such as cisplatin,which forms bifunctional DNA adducts,monofunctional Pt complexes interact with DNA in a different manner,forming only Pt‑G monoadducts[21‑23].Furthermore,most monofunctional Pt complexes under‑go hydrolysis at a slower rate(than classical bifunction‑al Pt drugs),and they are less reactive to thiol mole‑cules(e.g.,glutathione)in vivo,mainly due to their comparative chemical inertness and stronger hydropho‑bicity[24].Conclusively,monofunctional Ptcomplexes bind only one strand of DNA and might target cancer without causing auditory side effects,thus having more potential development and application significance than bifunctional drugs,including cisplatin.
Considering efforts to improve the anticancer activities of platinum‑based agents,the selection of non‑leaving ligands is especially important when designing and preparing monofunctional platinum com‑plexes.Groups containing nitrogen(N)donors are usu‑ally employed as leaving groups due to the thermody‑namic stability of the Pt—N bonds.Thus,many mono‑functional platinum complexes adopt a tridentate Pt‑NNN system.For instance,Liang′s group synthesized two monofunctional Ptcomplexes containing jatror‑rhizine derivative ligands([Pt(Jat1)Cl]Cl and[Pt(Jat2)Cl]Cl).Both of these compounds displayed remarkable anti‑tumor activity on HeLa cells(with IC50values of 15.01 and 1.00 nmol·L-1,respectively),and this anti‑tumor activity was achieved by targeting p53 and telom‑erase[25].Guo and co ‑workers developed a triphe‑nylphosphonium‑modified terpyridine platinumcom‑plex (TTP).TTP exhibited excellent cytotoxicity against cisplatin‑insensitive human ovarian cancer cells and preferential inhibition to mitochondrial TrxR[26].
In the present study,a novel binuclear monofunc‑tional Ptcomplex with a Pt‑NNN coordination mode,[Pt2(BPA‑TPA)Cl2]Cl2(Pt2‑BPA‑TPA,BPA‑TPA=2,6‑bis((bis(pyridin‑2‑ylmethyl)amino)methyl)pyridine),was synthesized(Scheme 1).Pt2‑BPA‑TPA was then characterized by NMR and high‑resolution mass spec‑troscopy(HRMS).X‑ray single crystal diffraction was also used to determine the structure of Pt2‑BPA‑TPA.Thereafter,thein vitroDNA cleavage activity of Pt2‑BPA‑TPA and its toxicity in the A549 cancer cell line were determined.Besides,we provided evidence for a credible molecular mechanism underlying its antitumor activities.Together,our results prove that Pt2‑BPA‑TPA is a promising candidate for novel anticancer therapy.

Scheme 1 Synthetic routes of ligand BPA‑TPA and complex Pt2‑BPA‑TPA
1 Experimental
1.1 Materials and physical measurements
All reagents were commercially obtained and,unless otherwise stated,were used without further puri‑fication.2,6‑Pyridinedimethanol was purchased from Nanjing Chemlin Co.,Ltd.Bis‑(2‑pyridylmethyl)amine(DPA)and K2PtCl4were obtained from Adamas Reagent Co.,Ltd.CCK‑8(cell counting kit‑8)was pur‑chased from Dojindo Molecular Technologies Inc.,Tokyo,Japan.
NMR spectra were measured on an AVANCE Ⅲ HD 400 MHz spectrometer.Electrospray ionization(ESI)mass spectra were obtained using an LCQ Fleet ThermoFisher mass spectrometer.Fluorescence spec‑tra and UV‑Vis absorption spectrum were determined on a Hitachi F‑4700 fluorescence spectrometer and VARIAN CARY 50 Conc UV‑visible spectrophotome‑ter,respectively.The crystal structure of the complex was determined on a Rigaku XtaLAB PRO Ⅱ single‑crystal diffractometer.
1.2 Synthesis of complex Pt2‑BPA‑TPA
2,6‑Bis(bromomethyl)pyridine was synthesized by previously reported methods[27].BPA ‑TPA was pre‑pared according to the reference[28].A 2,6‑bis(bromo‑methyl)pyridine(1.27 g,4.8 mmol)solution,excess K2CO3(1.59 g,11.5 mmol),and DPA(1.92 g,9.6 mmol)were added to CH3CN under N2atmosphere and stirred at 30 ℃ for 2 h.After completion of the reac‑tion,the mixture was filtered and the solvent was removed.The resulting yellow oil was extracted four times using boiling petroleum ether(b.p.60‑90 ℃)under vigorous stirring.BPA‑TPA was then precipitat‑ed from the gradually cooled petroleum solution as white crystals.The purified product(2.41 g)was obtained with a yield of 56% .1H NMR(400 MHz,CDCl3):δ8.51(d,J=4.8 Hz,4H),7.65‑7.57(m,9H),7.43(d,J=7.7 Hz,2H),7.14‑7.11(m,4H),3.91(s,8H),3.89(s,4H).13C NMR(100 MHz,CDCl3):δ159.32,158.61,149.05,136.87,136.45,123.00,122.00,121.17,60.20.HRMS in CH3OH(m/z):Calcd.[M+H]+502.272;Found:502.322.
K2PtCl4(410 mg,1 mmol)was first dissolved in 10 mL deionized water and then mixed with 10 mL metha‑nol containing BPA‑TPA(248 mg,0.5 mmol)in a round‑bottom flask.The resulting mixture was stirred in the dark for 48 h at room temperature.A beige precipitate was collected by centrifugation,and this was carefully washed with dichloromethane,ethanol,acetone,and anhydrous ether.The purified product of Pt2‑BPA‑TPA(52 mg)was obtained with a yield of 21% .1H NMR(400 MHz,DMSO‑d6):δ8.60(d,J=5.0 Hz,4H),8.20‑8.16(m,4H),7.74(d,J=7.9 Hz,4H),7.59(s,3H),7.55(t,J=6.4 Hz,4H),5.40(d,J=15.7 Hz,4H),5.06(d,J=15.8 Hz,4H),4.26(s,4H).HRMS in CH3CN(m/z):Calcd.[M-Cl]+998.095 7;Found:998.096 4.Calcd.[M-2Cl]2+481.065 2;Found:481.063 7.Calcd.[M-2Cl+6H2O]2+534.095 8;Found:534.092 3.
1.3 Single‑crystal structure determination
The crystal structure of Pt2‑BPA‑TPA was deter‑mined by the single‑crystal X‑ray diffraction method.Whitish block single crystals were obtained upon vapor diffusion of ethyl acetate into a solution of the complex in DMSO.Data for Pt2‑BPA‑TPA was collected on a hybrid pixel array detector(HPAD)at 293 K using graphite monochromatic CuKαradiation(λ=0.154 184 nm)in the multi‑scan mode.The structure was solved by direct methods using the SHELXS program of the SHELXTL package and refined using full‑matrix least‑squares methods with SHELXL(semi‑empirical absorp‑tion corrections were applied using the SADABS pro‑gram).SQUEEZE was adopted to reduce the effect orig‑inating from disordered solvent molecules.All hydro‑gen atoms were placed in a theoretical position.Crystal data and details of data collection and refinement for Pt2‑BPA‑TPA are summarized in Table 1.

Table 1 Crystal parameters and structure refinements for Pt2‑BPA‑TPA
CCDC:2115372.
1.4 DNA cleavage studies
DNA cleavage efficiency was assessed by agarose gel electrophoresis employing supercoiled pBR322 plasmid DNA.Briefly,supercoiled pBR322 plasmid DNA(200 ng)in 1×TAE buffer(Tris‑HCl‑EDTA)was treated with Pt2‑BPA‑TPA or BPA‑TPA(5‑20 μmol·L-1)in a final volume of 20 μL.All samples were incu‑bated at 37℃for 1.5 h.After treatment,loading buffer(3 μL)was added,and agarose gel electrophoresis was performed on a 1% gel(containing 1.0 μg·mL-1neutral red)at 100 V for 1 h in diluted 1×TAE buffer.The gel was then visualized by photographing intercalated neu‑tral red fluorescence under a UV illuminator.The cleavage efficiency was measured by determining the ability of the compound to excise supercoiled pBR322 DNA(SC)to its nicked circular(NC)and linear(L)forms.
1.5 CCK‑8 assay
The cytotoxicity of Pt2‑BPA‑TPA and cisplatin to human lung cancer A549 cells was measured using the CCK‑8 assay.CCK‑8 contains Dojindo′s highly water‑soluble tetrazolium salt,2‑(2‑methoxy‑4‑nitrophenyl)‑3‑(4‑nitrophenyl)‑5‑(2,4‑disulfophenyl)‑2H‑tetrazolium sodium salt(WST‑8),which produces a water‑soluble formazan dye upon reduction in the presence of an electron mediator.A549 cells were seeded in 96‑well plates at a density of 4×103cells per well and then incubated overnight.The adherent cells were then treated with different concentrations of drugs.After two days,10 μL of the CCK‑8 solution was added to each well of the plate,and the plate was incubated for 3 h in the incubator(37℃,φCO2=5% ).Finally,the OD450value(optical density at 450 nm)was measured in a microplate reader.
1.6 Western blot analysis
A549 cells(or HepG2 cells)were homogenized in lysis buffer(#9803,CST,USA),and total protein was isolated by centrifugation at 12 000 r·min-1for 15 min at 4℃.Total protein concentration in the lysates was quantified using a bicinchoninic acid(BCA)protein assay kit(A53225,Thermo Fisher Scientific,USA)according to the manufacturer′s instructions.The sam‑ples were then subjected to western blot analysis using specific antibodies againstα‑tubulin (GB11001,ServiceBio,Wuhan,China),cleaved caspase‑3(9661S,CST,USA),and p21(A1483,ABclonal,Wuhan,China)as described previously[29].
1.7 Statistical analysis
The data were analyzed by SPSS 19.0 software and presented as means±SD from at least three inde‑pendent experiments.All the statistical results were analyzed according to the Student′st‑testand Graphpad Prism 9.0(GraphPad Software Inc.,La Jolla,USA)was used to compare group means.A value ofp<0.05 represented statistically significant.
2 Results and discussion
2.1 NMR and HRMS characterization of Pt2‑BPA‑TPA
The structural characteristics of ligand BPA‑TPA and complex Pt2‑BPA‑TPA were elucidated by NMR and HRMS.The1H NMR spectra of Pt2‑BPA‑TPA and BPA‑TPA in DMSO‑d6are shown in Fig.1.In com‑parison with the spectrum of BPA‑TPA,proton signals in Pt2‑BPA ‑TPA were shifted significantly to the lower field,suggesting that Ptwas coordinated to the ligand,and thus providing evidence of the formation of a binuclear Ptcomplex in Pt2‑BPA‑TPA.The coor‑dination of Pt increased steric hindrance in the two pyr‑idyl groups,eliciting magnetic nonequivalence in the nearby methylene protons.Consequently,the H7 sin‑glet at 3.79 was split into two doublets at 5.4 and 5.1.The existence of Pt2‑BPA‑TPA in the CH3CN solution was also studied with HRMS.Them/zpeaks for[(Pt2‑BPA‑TPA)-Cl]+,[(Pt2‑BPA‑TPA)-2Cl]2+,and[(Pt2‑BPA‑TPA)-2Cl+6H2O]2+were observed at 998.096 4,481.063 7,and 534.092 3,respectively.The results indicate that complex Pt2‑BPA ‑TPA is stable in CH3CN.

Fig.1 1H NMR spectra of complex Pt2‑BPA‑TPA and the corresponding ligand BPA‑TPA
2.2 Crystal structure
As shown in Fig.2,binuclear Ptcomplex Pt2‑BPA‑TPA crystallizes in a monoclinic system with space groupP21/c(centrosymmetric space group).The two Ptions are symmetrically located in the center of the two pyridine ring arms which originate from DPA.Each Ption adopts an approximate planar quadrilat‑eral geometry associated with the[PtN3Cl]arrange‑ment,which is comprised of two pyridyl nitrogen atoms(for Pt1,N1 and N3;for Pt2,N4 and N6),a tertiary amine nitrogen atom(for Pt1,N2;for Pt2,N5),and a chloride atom(for Pt1,Cl1;for Pt2,Cl2).Using Pt1 as an example,the two diagonal positions are analogously occupied by N1 and N3 to yield Pt1—N1(0.202 7(14)nm)and Pt1—N3(0.203 9(14)nm).The dihedral angle between Cl1—Pt1—N3 and N1—Pt1—N2 is 2.166°.This dihedral angle deviates from the ideal value of 0°,implying a distorted square‑planar configuration of[PtN3Cl].Selected bond lengths and bond angles are listed in Table 2.
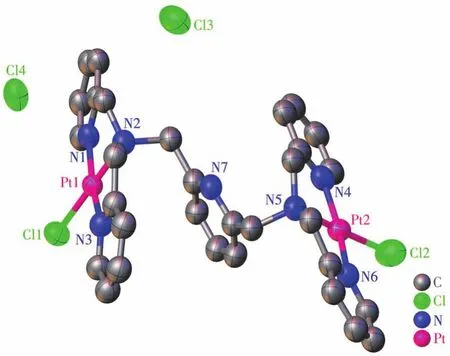
Fig.2 Crystal structure of Pt2‑BPA‑TPA with 50% thermal ellipsoids

Table 2 Selected bond lengths(nm)and angles(°)for Pt2‑BPA‑TPA
2.3 DNA cleavage activity
The main target of platinum anticancer agents is generally believed to be nuclear DNA.The mechanism of action of Ptcomplexes involves the formation of Pt‑DNA adducts that induce DNA damage by interfer‑ing with DNA transcription and replication,ultimately leading to cell death.To investigate the DNA cleavage activity of complex Pt2‑BPA‑TPA,we performed aga‑rose gel electrophoresis experiments using pBR322 plasmid DNA(1 μL,0.2 μg)in Tris‑HCl/NaCl(50 mmol·L-1,pH=7.2)buffer.A significant difference in the migration rates of the three types of DNA could be observed through gel electrophoresis.The migration rates can be expected to differ as follows:supercoiled form(Form Ⅰ,SC)>linear(Form Ⅲ,L)>nicked cir‑cular(Form Ⅱ,NC)[30].Fig.3 shows the results of gel electrophoretic separation of pBR322 plasmid DNA treated with increasing concentrations of Pt2‑BPA ‑TPA,and with the free ligand BPA‑TPA.Both Pt2‑BPA‑TPA and BPA‑TPA cleaved Form Ⅰ DNA into Form Ⅱ DNA and Form Ⅲ DNA in a concentration‑dependent manner.At the lowest concentration tested(5 μmol·L-1),supercoiled Form Ⅰ DNA was still observed,implying the complex′s weak nuclease activi‑ty at such a faint dose.Surprisingly,at 10 μmol·L-1concentration,Pt2‑BPA‑TPA and BPA‑TPA exhibited efficient cleavage activity;accordingly,Form Ⅰ was relaxed to generate Form Ⅱ and Ⅲ,suggesting that double‑stranded DNA was thoroughly cleaved.Under the same concentration conditions,the DNA cleavage activity of the binuclear Ptcomplex was not signifi‑cantly higher than that of the ligand.We hypothesize that the introduction of two Ptions does not signifi‑cantly change the spatial structure of the ligand.

Fig.3 Agarose gel electrophoresis diagrams for cleavage of pBR322 plasmid DNA induced by complex Pt2‑BPA‑TPA and ligand BPA‑TPA
2.4 In vitro cytotoxicity

Fig.4 Dose‑dependent curves showing the cytotoxicity of cisplatin and complex Pt2‑BPA‑TPA in A549 cells
2.5 Expression of apoptosis‑related proteins
Since complex Pt2‑BPA‑TPA exhibited increased anticancer activity(compared to cisplatin),we further explored its anticancer mechanism in greater detail.p21 protein and caspase family proteins are both known to be key regulators of the apoptotic pathway.If p21 is produced in excess,the affected cellproceeds to apoptosis[33].Caspase‑3 is an important“execution‑er”of this apoptotic process[18,34].To gain an insight into the molecular mechanism of cell death triggered by Pt2‑BPA ‑TPA,we used western blot assays to detect p21 and caspase‑3 protein levels in HepG2,A549 cells,and rat myocardial(H9c2)cells.All three cell types were treated with Pt2‑BPA‑TPA at concen‑trations of 10 or 20 μmol·L-1.For comparison,these cells were also treated with cisplatin at 1 and 5 μmol·L-1.As shown in Fig.5,p21 and cleaved‑caspase‑3 protein levels in HepG2 and A549 cells were markedly increased by Pt2‑BPA‑TPA treatment.In A549 cells,a six‑fold increase in the p21 protein level was observed,and this was significantly higher than that observed in HepG2 cells.No significant increase in either p21 protein or cleaved‑caspase‑3 protein levels was observed in normal H9c2 cells.These data provide solid evidence that Pt2‑BPA ‑TPA induces critical nuclear DNA damage,subsequently inducing an upreg‑ulation in the protein levels of important downstream apoptosis‑related proteins,including p21 and cleaved‑caspase‑3.

Fig.5 Expression of p21 and cleaved‑caspase‑3(C‑casp 3)in HepG2,A549,and H9c2 cells after incubation with cisplatin(Cis‑Pt)and Pt2‑BPA‑TPA for 24 h,where α‑tubulin used as an internal control(CON)
3 Conclusions
In summary,we have successfully prepared a new binuclear monofunctional Ptcomplex,Pt2‑BPA‑TPA,based on the Pt‑NNN coordination pattern and employing polypyridyl as the non‑leaving group.Single‑crystal X‑ray diffraction analysis reveals that Pt2‑BPA‑TPA belongs to the monoclinic system with space groupP21/c.The cleavage activity of Pt2‑BPA‑TPA towards supercoiled pBR322 plasmid DNA was also studied by agarose gel electrophoresis,demonstrating that Pt2‑BPA ‑TPA could effectively cleave Form Ⅰ DNA to Form Ⅱ DNA and Form Ⅲ DNA.CCK‑8 anal‑ysis revealed that Pt2‑BPA ‑TPA could inhibit the growth of A549 cells and that Pt2‑BPA‑TPA demon‑strated higher cytotoxicity than cisplatin after 48 h incubation.Moreover,western blot analyses provided evidence that the anticancer mechanism of Pt2‑BPA‑TPA involves the upregulation of protein levels of important downstream apoptosis‑related proteins,including p21 and cleaved‑caspase‑3.Generally,Pt2‑BPA ‑TPA,a refinedly synthesized binuclear mono‑functional Ptcomplex,is a promising candidate for novel anticancer therapy.
Declaration of competing interest:All authors of this paper declare that they have no competing interests.
Acknowledgments:This project was supported by the Educational Commission of Hubei Province of China(Grant No.D20212802),the Natural Science Foundation of Hubei Province(Grant No.2021CFB439),and the Scientific Projects of Health Commission of Hubei Province(Grant No.WJ2019Q022).
