Comparative acetylome analysis reveals the potential mechanism of high fat diet function in allergic disease
2023-01-23YnnSunNingLiuHuihuiWngTiqiQuFzhengRenYixunLi
Ynn Sun, Ning Liu, Huihui Wng, Tiqi Qu, Fzheng Ren, Yixun Li,*
a Key Laboratory of Precision Nutrition and Food Quality, Department of Nutrition and Health, China Agricultural University, Beijing 100193, China
b College of Food Science and Engineering, Gansu Agricultural University, Lanzhou 730070, China
Keywords:Allergic disease High fat diet Skeletal muscle Acetylome
A B S T R A C T Modern technological lifestyles promote allergic diseases, especially food allergies. The underlying molecular mechanisms remain to be uncovered. Protein acetylation is one of the most important post-translational modif ications, and it is involved in regulating multiple body metabolic processes. This study aimed to clarify the effects of a high-fat diet (HFD) on allergy risk and the underlying mechanisms. Four-week-old male C57BL/6J mice were randomly divided into two groups and fed a normal fat diet (NFD) or HFD for 24 weeks.Then, serum lipids were measured, and skeletal muscle was collected for acetylome analysis. Compared with the findings in the NFD group, HFD-fed mice were obese and hyperlipidemic. Acetylome analysis also revealed 32 differentially expressed proteins between the HFD and NFD groups. Among these, eight acetylated proteins were upregulated in the HFD group. In addition, 13 and 11 proteins were acetylated only in the HFD group and NFD group, respectively. These proteins were mainly involved in regulating energy metabolism and mitochondrial function. This study provides information regarding the underlying molecular mechanisms by which HFD promotes allergy.
1. Introduction
The incidence of obesity and food allergies has been increasing in recent decades. Studies suggest that modern technological lifestyles(including the standard Western diet rich in fat and sugar) promote these diseases [1,2]. Hussain et al. [3] revealed that HFD-induced obesity increases susceptibility to food allergies. Although accumulated experimental data have indicated that high-fat diet (HFD)-induced obesity has a close relationship with food allergies and inflammation,the molecular mechanism by which HFD consumption promotes food allergies remains to be clarif ied [4-6]. Recent studies indicated that mitochondria dysfunction and dysregulated gut microbiota play important roles in the development of food allergies [7-13]. Free fatty acids (FFAs) are the primary components of HFDs. Increasing serum FFAs concentrations decrease the expression of intestinal tight junction proteins [3]. However, excessive FFAs content causes allergens to enter the immune system through increasing lipopolysaccharide (LPS) levels [3]. Increasing FFAs levels activate the nuclear factor-κB pathway, which regulates the expression of pro-inflammatory genes involved in the production of cytokines,such as IL-1β and TNF-α [14]. FFAs mainly regulate oxidative phosphorylation in mitochondria. Excessive FFAs content results in mitochondrial overload and excessive reactive oxygen species(ROS) production, thereby aggravating inflammation [15]. Allergic diseases such as asthma, atopic dermatitis, and allergic rhinitis are accompanied by mitochondrial dysfunction, increasing ROS levels,and systemic inflammation [16,17]. A hypothesis suggested that mitochondrial connects allergies, inflammatory predisposition, and metabolic diseases.
Proteomics is an effective strategy for discovering new pathways. Proteins are responsible for intracellular signaling,and they are the targets for disease treatment. Changes in protein structure and interactions contribute to the underlying mechanism of many diseases, including allergies and related diseases [18].Post-translational modifications (PTM) of proteins regulate their cellular localization, activity, and interactions with other proteins,thereby playing a pivotal role in functioning. Lysine acetylation is catalyzed by acetyltransferases (KATs) and deacetylases (KDACs).This widespread and reversible PTM is involved in multiple cellular processes in eukaryotic cells. First identified as a histone modification,a recent study suggested that approximately 4 500 proteins (including transcription factors, other nuclear proteins, and cytoplasmic proteins)can be acetylated at approximately 15 000 acetylation sites [19,20].Whereas non-histone protein acetylation is poorly understood,recent findings suggested that most enzymes that regulate glycolysis,gluconeogenesis, fatty acid metabolism, and the tricarboxylic cycle(TCA) are acetylated in human liver tissue [21]. These studies indicated that lysine acetylation of proteins plays an important role in the regulation of glucolipid metabolism. Because acetylation is reversible, further investigation of protein acetylation might uncover novel therapeutic targets for the treatment of allergies and related chronic metabolism diseases.
β-Oxidation of FFAs in mitochondria requires several metabolic enzymes that require acetylation to exert their biological effects.This study examined the relationship between HFD-induced obesity and allergy. Therefore, acetylome analysis of skeletal muscle with mice fed a normal fat diet (NFD) or HFD was performed, and the findings revealed several new acetylated proteins acetylation (and the sites of acetylation) regulating inflammation and allergy signaling pathways.
2. Materials and methods
2.1 Animals
Four-week-old male wild-type C57BL/6J mice (n= 30) were purchased from Beijing HFK Bioscience Co., Ltd. After 1-week adaptation period, 6 mice were removed because they were underweight (bodyweight < 15 g). Then, the mice were randomly assigned to receive a control diet (NFD, 10% calories from fat)or HFD (60% calories from fat) for 24 weeks. Mice were housed in a temperature-controlled ((22 ± 2) °C) room with a 12 h/12 h light/dark cycle and (55 ± 10)% relative humidity, and they were permitted free access to food and water. Mice were fasted for 12 h and then sacrificed using diethyl ether. Serum was collected by centrifugation at 3 170 ×gfor 15 min at 4 °C and stored at -80 °C for further analysis. Serum lipids were measured by the Core Clinical Laboratory of the No.3 Hospital of Beijing University using an automatic biochemical analyzer (Hitachi 7600-020). Skeletal muscle was removed, rinsed with PBS, and stored at -80 °C. This study was conducted in strict accordance with the recommendations in the Guide for the Institutional Animal Care and Ethics Committee of the China Agricultural University (CAU20170430-4).
2.2 Acetylome analysis
2.2.1 Sample preparation
Skeletal muscle was frozen in liquid nitrogen and ground with a pestle and mortar. Urea was added to the powder, and the lysate was sonicated on ice. After centrifugation at 18 000 ×gfor 10 min,the supernatant was quantified using Bradford reagent. Then,the proteins were separated on a 12.5% sodium dodecyl sulfate polyacrylamide gel electropheresis (SDS-PAGE) gel (constant current 15 mA, 60 min). Each sample was mixed with 10 mmol/L dithiothreitol (DTT) buffer, IAA was added, and the samples were incubated for 30 min in darkness. Next, trypsin was added to digestion for 18 h at 37 °C. The peptides of each sample were desalted on C18Cartridges (Waters WAT051910).
2.2.2 Enrichment of acetylated peptides
Samples were reconstituted in IAP buffer, and enrichment of acetylated peptides was performed using Anti-Ac-K antibody beads(PTMScan Acetyl Lysine Motif (Ac-K) Kit, Cell Signal Technology).
2.2.3 LC-MS/MS analysis
MS data were acquired using a data-dependent top 10 method by dynamically choosing the most abundant precursor ions from the survey scan (350-1 800m/z) for HCD fragmentation. Survey scans were acquired at a resolution of 70 000 atm/z200, and the resolution for HCD spectra was set to 17 500 atm/z200.
2.3 Data analysis
Statistical analysis was performed using SPSS statistical software(version 21.0). The statistical significance of the data was assessed using Student’st-test.P< 0.05 indicated statistical significance.
3. Results
3.1 Differentially expressed acetylated protein between the two groups
The basic information on experimental animals is presented in Supplemental Table 1. Bodyweight was increased by 47%in the HFD group. The concentrations of fasting glucose, total cholesterol, total triglycerides, low-density lipoprotein cholesterol,and high-density lipoprotein cholesterol were 2.8-, 2.0-, 1.4-,2.8-, and 1.73-fold higher, respectively, in the HFD group than in the NFD group (Supplemental Table 1). Pro-inflammatory cytokine levels were significantly increased in the HFD group(Supplemental Fig. 1). The changes in body weight and serum lipid levels suggested that mice in the HFD group were obese and hyperlipidemic. Acetylome analysis uncovered 4 817 acetylated peptides, including 617 acetylated proteins. In total, 32 acetylated proteins exhibited significantly different abundance between the HFD and NFD groups. Among these proteins, eight acetylated proteins were upregulated in the HFD group (Table 1). In addition,13 and 11 proteins were acetylated only in the HFD and NFD groups, respectively (Tables 2-3).
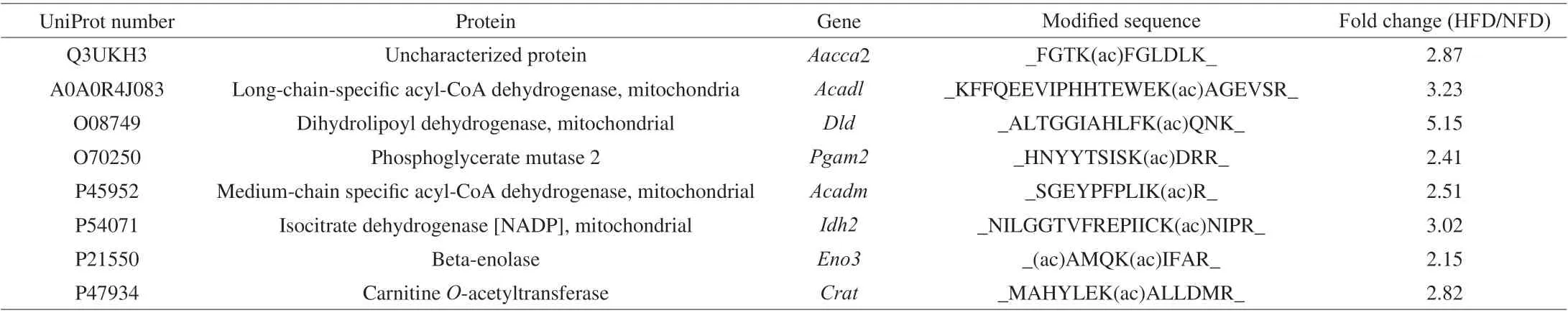
Table 1 Differentially expressed acetylated proteins induced by HFD feeding.
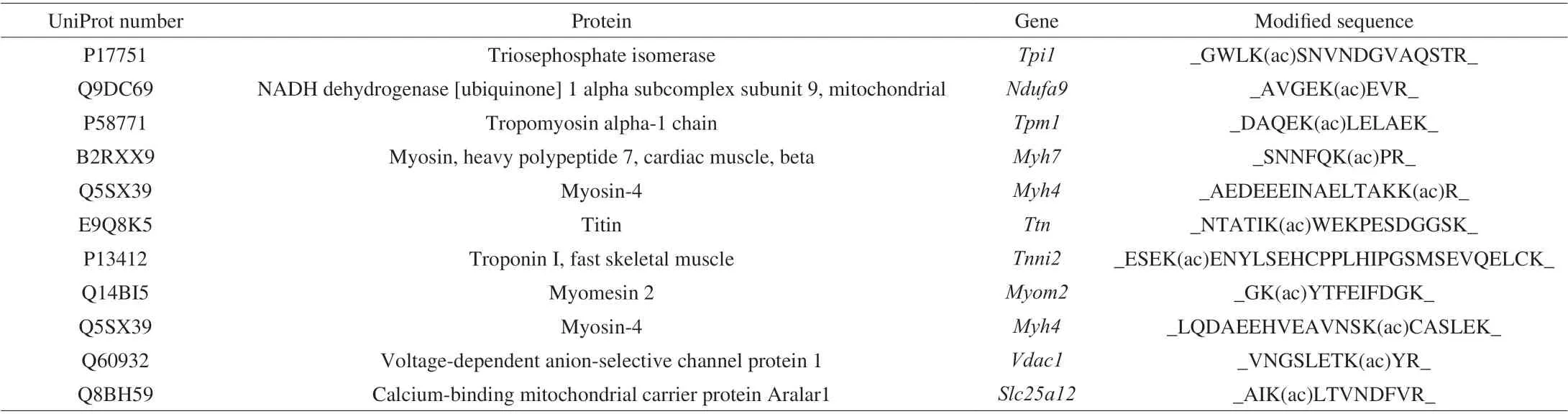
Table 2 Proteins acetylated only in the normal fat diet group.
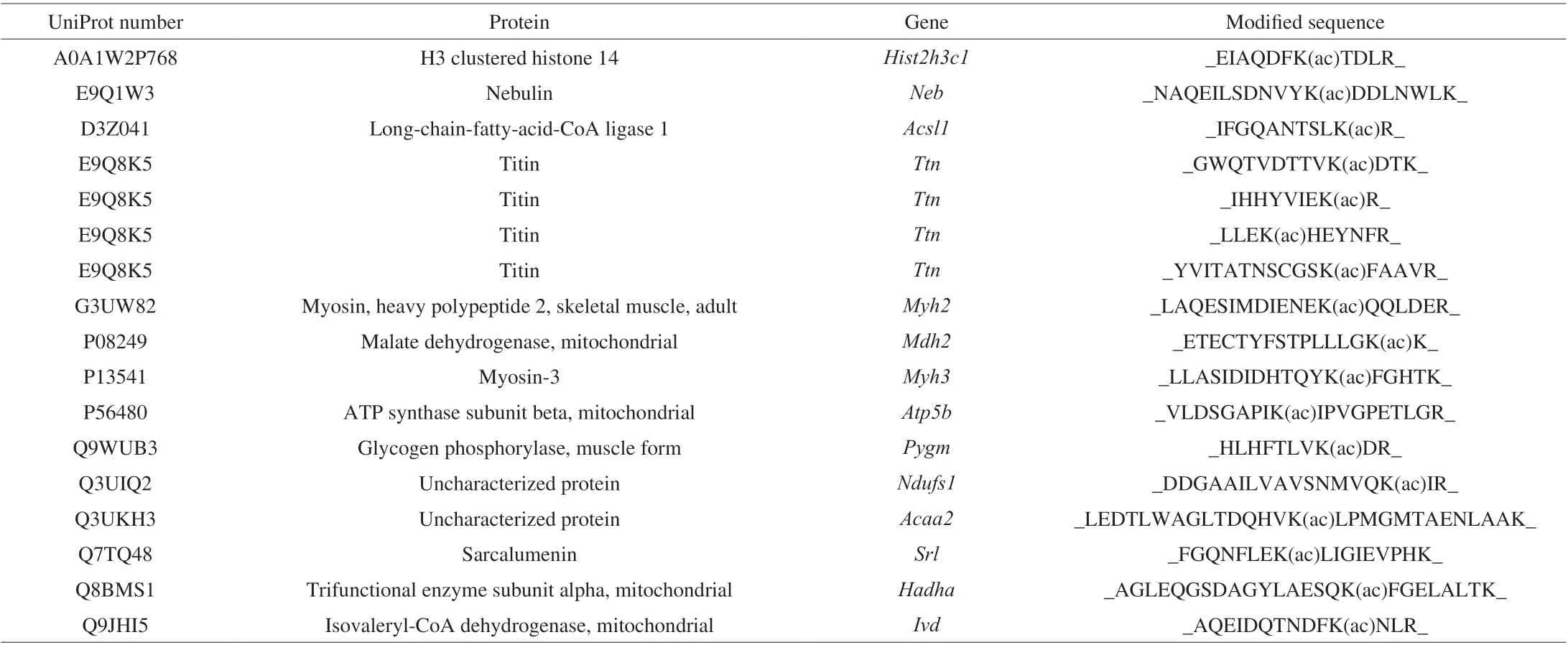
Table 3 Proteins that were only acetylated in the high-fat diet group.
As presented in the tables, the acetylated proteins were mostly involved in regulating glucolipid metabolism and mitochondrial function. The results indicated that HFD feeding regulated the acetylation of enzymes involved in energy metabolism.
3.2 Functional enrichment analysis
In total, 8 acetylated muscle proteins had significantly different abundance between the NFD and HFD groups. In Fig. 1, the 8 acetylated peptides were marked from blue to red to abundance. A redder color indicates a higher acetylation level, whereas a bluer color indicates a lower level of acetylation. This result revealed that these eight proteins had significantly higher acetylation levels in the HFD group.
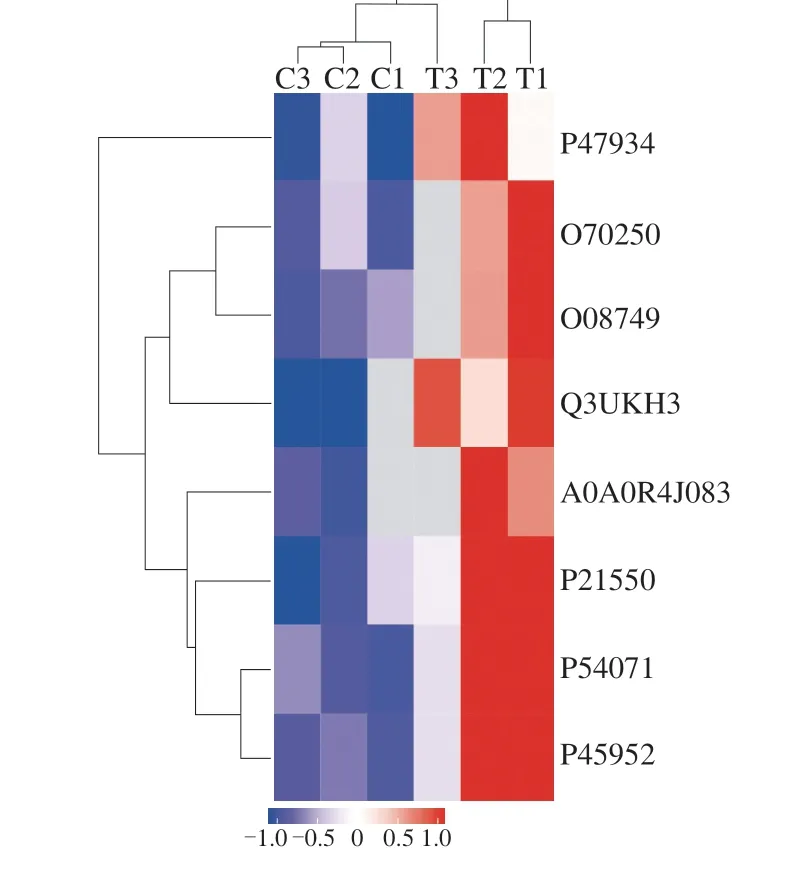
Fig. 1 Clustering of differentially expressed acetylated peptides in the NFD and HFD groups. C1, C2, and C3 are the three samples in the NFD group, and T1, T2, and T3 are the three samples in the HFD group.
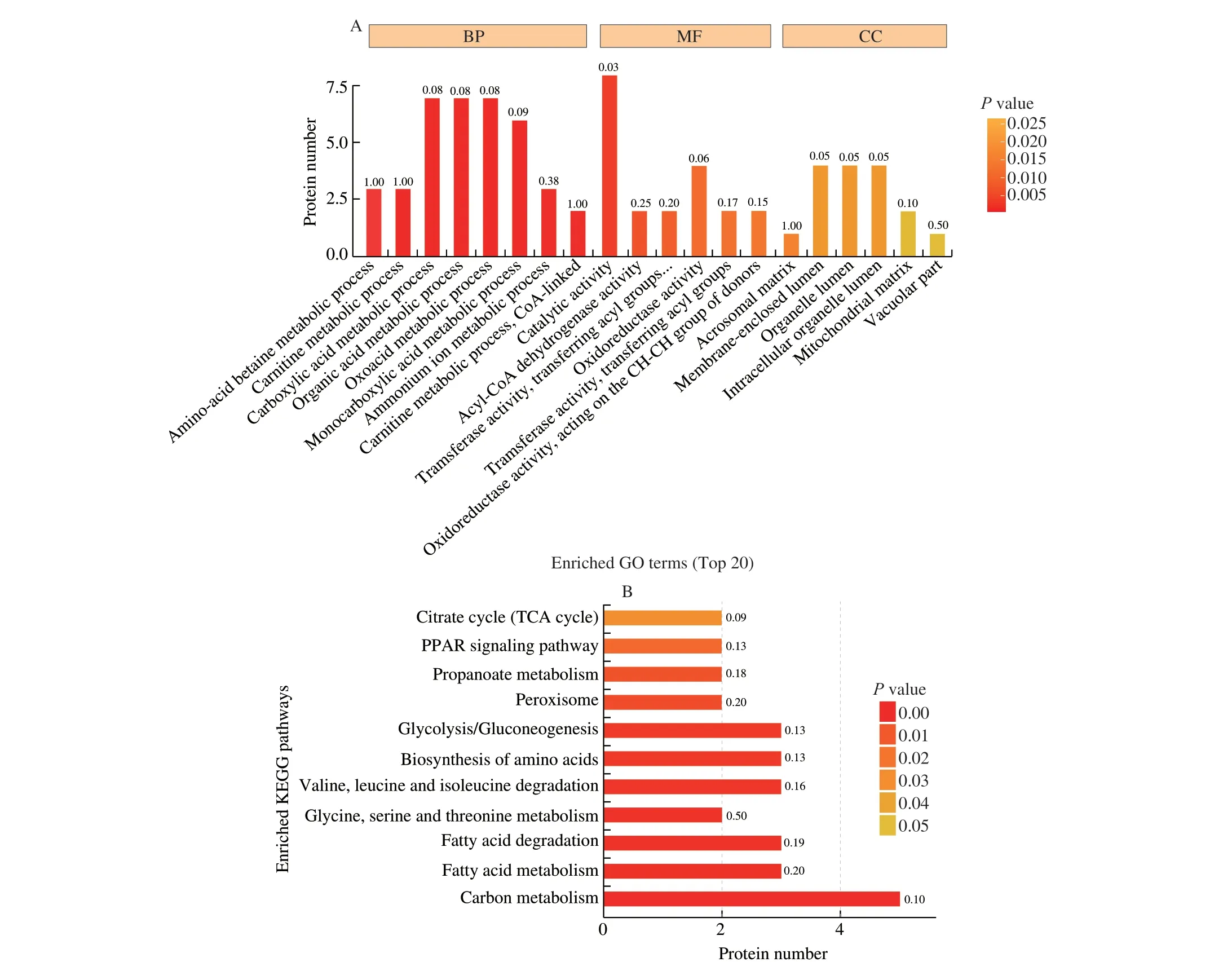
Fig. 2 GO and KEGG pathway enrichment analyses of the identified acetylated proteins. (A) GO enrichment analysis of the upregulated acetylated proteins in the HFD group. (B) KEGG pathway enrichment analysis of the upregulated acetylated proteins in the HFD group. BP, biological process;MF, molecular function; CC, cellular compartment.
3.3 Functional enrichment analysis
Functional enrichment analysis of differentially expressed acetylated proteins in the two groups was conducted. In the gene ontology (GO) analysis of biological processes, the acetylated proteins were enriched in amino-acid betaine metabolic, carnitine metabolic,carboxylic acid metabolic, organic acid metabolic, oxoacid metabolic,monocarboxylic acid metabolic, ammonium ion metabolic, and carnitine metabolic (CoA-linked) processes. In the GO analysis of molecular functions, the acetylated proteins were enriched in catalytic activity, oxidoreductase activity, and acyl-CoA dehydrogenase activity.In the GO of cellular compartments, the acetylated proteins were enriched in the organelle lumen, mitochondrial matrix, and membraneenclosed lumen. Kyoto Encyclopedia of Genes and Genomes (KEGG)pathway enrichment revealed that the acetylated proteins were enriched in several pathways including the citrate cycle (TCA cycle), peroxisome proliferators-activated receptor (PPAR) signaling pathway, glycolysis/gluconeogenesis, fatty acid metabolism, and carbon metabolism.
As presented in Fig. 2, GO and KEGG pathway enrichment provided evidence of the role of acetylation in energy metabolism and mitochondrial function.
4. Discussion
The incidence of allergic diseases such as food allergies is rising globally, but the mechanisms supporting their development are unclear. HFD feeding induced inflammation, obesity, and allergic diseases. Previous studies on the mechanism by which HFD feeding promotes food allergies mainly focused on the dysregulation of gut microbiota and intestinal permeability.HFD feeding leads to the accumulation of LPS, and triggered inflammatory responses [22]. HFD feeding can reduce the expression of epithelial tight junction proteins, such as occludin and ZO-1, and these changes result in increased intestinal permeability [23,24].In this study, we revealed that mitochondrial dysfunction and dysregulated energy metabolism also play an important role in the development of allergic diseases.
HFD feeding significantly promoted the acetylation of phosphoglycerate mutase 2 and beta-enolase, which are key enzymes of glycolysis. Acetylation of glycogen phosphorylase was also increased by HFD feeding, and this acetylated protein plays an important role in glycogenolysis. HFD feeding might regulate the TCA cycle by increasing the acetylation of carnitineO-acetyltransferase and long-chain fatty acid CoA ligase 1. CarnitineO-acetyltransferase is located within the mitochondrial matrix, and it is involved in fatty acid oxidation [25]. Studies indicated that the deletion of carnitineO-acetyltransferase aggravated metabolic dysregulation in obese mice and induced the accumulation of longchain acyl-carnitines (an indicator of incompleteβ-oxidation) [26].Long-chain fatty acid CoA ligase 1 is required for the activation of long-chain fatty acids. Increased long-chain fatty acid CoA ligase 1 expression in Schwann cells can protect mitochondrial function and decrease proton leak [27]. The increased acetylation of carnitineO-acetyltransferase and long-chain fatty acid CoA ligase 1 might inhibit their catalyze activation, leading to a decrease inβ-oxidation.
As demonstrated by Skop et al. [28] , the mRNA expression of genes that regulate mitochondrial function and energy metabolism,including genes involved in electron transport (Nufsd1), ATP synthesis (ATP5b), fatty acid metabolism (Acadl), and the TCA cycle (Mdh2), were differentially expressed in preadipocytes and differentiated adipocytes. All of these genes had higher levels of acetylation in the HFD group. This result indicated that HFD feeding both regulated mitochondrial function by affecting the mRNA expression of enzymes and increased the acetylation of these enzymes. Studies demonstrated that ATP synthase subunit beta might participate in mitochondrial dynamics and regulate mitochondrial fission and fusion [29,30]. Voltage-dependent anion-selective channel protein 1 (VDAC1) is located at the outer mitochondrial membrane,and it plays an important role in regulating mitochondrial metabolism and functions. VDAC1 deficiency can lead to embryonic lethality and decrease respiratory capacity [31]. VDAC1 also participates in cross-talk between the endoplasmic reticulum and mitochondrial Ca2+homeostasis exchange [32]. VDAC1 also regulates mitochondrial Ca2+homeostasis and oxidative phosphorylation by transporting Ca2+[33]. VDAC1 was only acetylated in the NFD group. These results indicate that the acetylation of VDAC1 regulates its biological function in mitochondria, contributing to mitochondrial homeostasis.NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 9(Ndufa9) is a Q-module subunit that is a component of mitochondrial respiratory chain complex I. Its deficiency induces complex I disease,and its mutations cause complex I assembly defects [34,35]. In addition, acetylated Ndufa9 was only detected in the NFD group. We hypothesized that the acetylation of VDAC1 may promote complex I assembly and stability.
In this study, we attempted to clarify the molecular mechanism by which HFD feeding promotes allergic diseases via acetylome analysis.Our results indicated that HFD feeding promotes allergic diseases possibly by acetylating mitochondrial proteins and energy enzymes.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
This research was funded by the 111 project from the Education Ministry of China (B18053). We thank Joe Barber Jr., from Liwen Bianji (Edanz) for editing the English text of a draft of this manuscript.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://doi.org/10.1016/j.fshw.2022.09.019.
杂志排行
食品科学与人类健康(英文)的其它文章
- The role of probiotics in prevention and treatment of food allergy
- Roles of fermented plant-, dairy- and meat-based foods in the modulation of allergic responses
- The role of gut microbiota and its metabolites short-chain fatty acids in food allergy
- Association of nutrients intake during pregnancy with the risk of allergic disease in offspring: a meta-analysis of prospective cohort studies
- Purif ication and immunoglobulin E epitopes identif ication of low molecular weight glutenin: an allergen in Chinese wheat
- Determination of egg and milk allergen in food products by liquid chromatography-tandem mass spectrometry based on signature peptides and isotope-labeled internal standard