Mechanism underlying the effect of Liujunzi decoction (六君子汤) on advanced-stage non-small cell lung cancer in patients after first-line chemotherapy
2022-11-17XINXiaoliWANGGuodongHANRuJIANGYiLIUChangLIULingshuangXUZhenye
XIN Xiaoli,WANG Guodong,HAN Ru,JIANG Yi,LIU Chang,LIU Lingshuang,XU Zhenye
XIN Xiaoli,JIANG Yi,LIU Chang,LIU Lingshuang,XU Zhenye,Department of Oncology,Longhua Hospital affiliated to Shanghai University of Traditional Chinese Medicine,Shanghai 200032,China
WANG Guodong,Department of Orthopedics,Longhua Hospital affiliated to Shanghai University of Traditional Chinese Medicine,Shanghai 20032,China
HAN Ru,Department of human resources,Shandong Academy of Occupational Health and Occupational Medicine,Jinan 250000,China
Abstract OBJECTIVE: To further clarify the anticancer mechanisms of Liujunzi decoction (六君子汤) and provide possible targets for the treatment of advanced-stage nonsmall cell lung cancer (NSCLC) by re-analyzing differential gene expression profile of peripheral blood mononuclear cells (PBMCs) from Liujunzi decoctiontreated NSCLC patients receiving first-line chemotherapy.METHODS: The PBMC gene expression microarray data set GSE61926 was retrieved from a high throughput gene expression database.Differentially expressed genes(DEGs) were screened by paired sample t-test and the multiple ratio method.Gene ontology and Kyoto encyclopedia of genes and genomes (KEGG) pathway analyses were performed using the DAVID database.The protein–protein interaction (PPI) network was constructed using interaction gene library retrieval tools and Cytoscape software.RESULTS: A total of 162 DEGs were identified,with 67 upregulated genes and 95 downregulated genes.The functional distribution of Gene Oncology (GO) genes showed that DEGs were mostly concentrated in extracellular regions,calcium ion binding,and transcriptase activity.KEGG pathway analysis showed that cytokine–cytokine receptor interactions were significantly enriched.PPI network analysis screened out the top 10 central protein-coding genes with the highest nodal degree: IL2,PIWIL4,DICER1,PIWIL2,SAA1,XCL1,IL22RA1,ARHGAP11A,DCP1A,and GDNF.Among them,the central protein-coding gene with the highest node degree was IL2.In addition,the central protein-coding genes with high node degrees and high molecular complex detection (MCODE) scores were PIWIL4,DICER1,PIWIL2,and DCP1A,all of which are related to tumor development.CONCLUSIONS: One signaling pathway and 10 central protein-coding genes related to anticancer mechanisms were screened by re-analysis of GSE61926 data.IL2,PIWIL4,DICER1,PIWIL2,and DCP1A may have important roles in the mechanism of Liujunzi decoction treatment against NSCLC.Our results suggest that the anticancer mechanism of Liujunzi decoction may berelated to gene silencing by RNA and the biological processes of piwi-interacting RNA and other small RNAs.
Keywords: carcinoma,non-small-cell lung;antineoplastic agents;computational biology;RNA,small interfering;Liujunzi decoction
1.INTRODUCTION
Non-small cell lung cancer (NSCLC) mainly includes adenocarcinoma and squamous cell carcinoma.Approximately 1.8 million people are diagnosed with lung cancer and 1.6 million die from it each year.The 5-year survival rates of lung cancer range from 4% to 17%,depending on the cancer staging and site.1NSCLC accounts for 80%-85% of lung cancers,and most NSCLC patients are first diagnosed at the middle and advanced stages,with a 5-year survival rate of 18%.2,3Currently,most NSCLC patients are treated with chemoradiotherapy or targeted therapy.However,because of such factors as the emergence of drug resistance,age-related intolerance,and a lack of sensitivity to chemoradiotherapy,NSCLC tends to metastasize and affect prognosis.Therefore,it is urgent to implement effective treatment and prognostic monitoring for NSCLC patients.Liujunzi decoction (六君子汤,Rikkunshito) is a compound decoction consisting of Dangshen (Radix Codonopsis),Baizhu(Rhizoma Atractylodis Macrocephalae),Fuling (Poria),Gancao (Radix Glycyrrhizae),Gouqizi (Fructus Lycii),Banxia (Rhizoma Pinelliae),Shengjiang (Rhizoma Zingiberis Recens),and Dazao (Fructus Jujubae),and originates from ancient China.It has high application value in the adjuvant treatment of anorexia and digestive tract cancer,and in the reduction of side effects from chemoradiotherapy.4-6Chenet al7used Liujunzi decoction to treat NSCLC patients after first-line chemotherapy,and PBMCs from the patients were collected for mRNA high-throughput sequencing.They found that Liujunzi decoction affected the blood neutrophil count by inhibiting neutrophil apoptosis and enhancing the transendothelial migration and adhesion of neutrophils,as confirming that anticancer immunity is conferred by Liujunzi decoction.In view of the complexity of cancer pathogenesis,we re-analyzed the data set to further clarify the anticancer mechanisms of Liujunzi decoction to provide possible targets for the treatment of advanced-stage NSCLC.
2.METHODS
2.1.High-throughput sequencing data acquisition
The data set GSE61926 was retrieved and downloaded from the Gene Expression Omnibus database of the National Center for Biotechnology Information.The platform was the Illumina humanRef-8 v2.0 expression beadchip.All 17 advanced NSCLC patients received a maximum of six courses of chemotherapy with cisplatin and gemcitabine in a 28-day cycle as first-line treatment.They were randomly assigned to receive Liujunzi decoction treatment (n=9) or dry powder placebo (n=8).The primary endpoint was the treatment response and the categories of the scales of anorexia,nausea,vomiting,and fatigue;secondary endpoints included the hematological effect and whole genome gene expression changes.7Gene expression in PBMCs was systematically analyzed by whole genome expression beadchip.Considering that only differentially expressed gene(DEG) screening and Gene Oncology (GO) analysis were performed in the original citation,the data set was retrieved for re-analysis in this study to investigate the anticancer mechanism.
2.2.Identification of DEGs
After robust multi-array average background correction,the data set GSE61926 was summarized and normalized to obtain single gene expression data.The online microarray data analysis software Morpheus(http://software.broadinstitute.org/morpheus) was used for the identification of DEGs by paired samplet-test.DEGs satisfying |lgFC| > 1.00 andP< 0.05 were screened out.
2.3.Gene oncology and pathway enrichment analysis
DEGs were imported into the online database the Database for Annotation,Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov) to perform GO and KEGG pathway analyses,andP-values of < 0.05 were considered statistically significant.Functional genes and critical signaling pathways were screened in this way.
2.4.Protein-protein interaction (PPI) analysis
DEGs were converted to official gene names using the DAVID network database and were imported to the Search Tool for the Retrieval of Interacting Genes(STRING) 10.0 database (http://string-db.org) to generate the network data set.Cytoscape software was used to apply PPI enrichment analysis and the Molecular Complex Detection (MCODE) algorithm to identify densely connected network components.Threshold set for the MCODE algorithm is that Degree Cutoff=2,Node density cutoff=0.1,Node Score Cutoff=0.2,KCore=2,Max.Depth=100.Among the 10 proteins with the highest node degree enriched in PPI analysis,the proteins enriched by the MCODE algorithm were selected and imported into the STRING 10.0 database for further GO and KEGG pathway analyses.
3.RESULTS
3.1.Identification of DEGs
DEG identification showed that 162 therapy-related DEGs were selected,among which 67 genes were upregulated and 95 genes were downregulated.A DEG expression heatmap with the most significant changes in gene expression level is shown in Figure 1.Upregulated genes are marked in red and downregulated genes in blue.
3.2.GO enrichment results
GO enrichment includes the analysis of biological process (BP),cellular components (CC),and molecular function (MF).The first five terms of BP categories with the lowestP-value were shown in Table 1.The CC enrichment results withP< 0.05 were shown in Table 2,which are associated with tumorigenesis.The MF enrichment results withP<0.05 were shown in Table 3.
3.3.KEGG pathway enrichment analysis
A total of one pathway met the threshold requirement ofP <0.05 and was significantly enriched,which was a signaling pathway related to the intervention mechanism of Liujunzi Decoction on advanced NSCLC patients after first-line chemotherapy: Cytokine-Cytokine receptor interaction signaling pathway,as shown in Table 4.By KEGG enrichment analysis,only the cytokine -cytokine receptor interaction pathway was significantly enriched,and the enrichment of DEGs is shown in Figure 2.The genes marked with red arrows are enriched DEGs,and the direction of the arrows represent their upregulation and downregulation.
3.4.PPI enrichment analysis
The PPI network includes nodes representing proteins and edges representing proteins interacting with each other.The top 10 proteins with the highest node degrees were shown in Table 5,among which IL2 ranked highest.The MCODE algorithm showed that three clusters were screened out,as shown in Figure 3.In addition to IL2,which had the highest node-degree,genes with high node degrees and MCODE scores included PIWIL4,DICER1,PIWIL2,and DCP1A,as highlighted in Figure 4.

Figure 2 Enrichment of differentially expressed genes in the cytokine-cytokine receptor interaction signaling pathway

Table 1 GO enrichment analysis (BP)

Table 2 GO enrichment analysis (CC)

Table 3 GO enrichment analysis (MF)

Table 4 KEGG pathway enrichment analysis

Figure 1 DEG expression heatmap (partial)
The MCODE algorithm and PPI analysis showed that there were proteins with relatively low node degrees in cluster B and C,while the four proteins in cluster A all had high node degrees.These proteins in cluster A were imported into the STRING 10.0 database for further analyses by GO and KEGG enrichment.Gene expression and RNA metabolism were enriched in GO analysis,asshown in Table 6.KEGG pathway analysis showed that gene silencing by RNA (HSA- 211000;Figure 5) and piwi-interacting RNA (piRNA) biogenesis (HSA-5601884,Table 7) were enriched.Therefore,the anticancer mechanism of Liujunzi decoction might be related to gene silencing by RNA in which piRNA was involved.

Table 7 KEGG enrichment analysis by STRING 10.0 (PIWIL4,DICER1,PIWIL2,and DCP1A)

Figure 5 Gene Silencing by RNA

Table 6 GO enrichment analysis by STRING 10.0 (PIWIL4,DICER1,PIWIL2,and DCP1A)
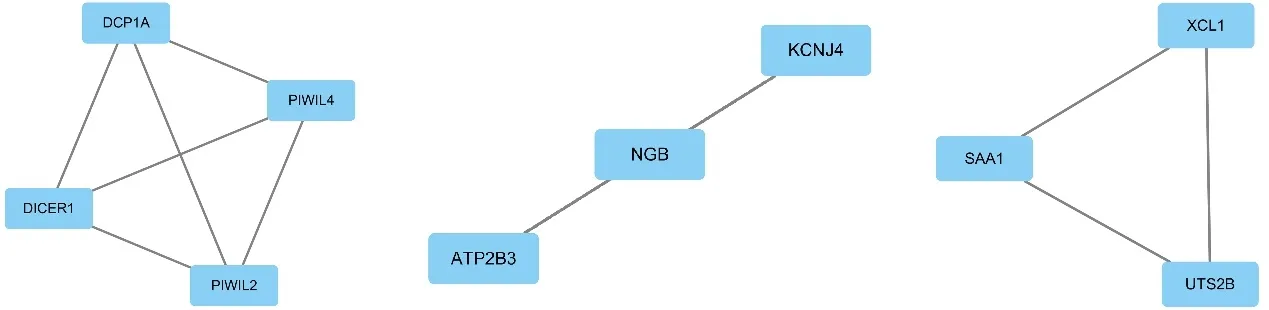
Figure 3 molecular complex detection clusters
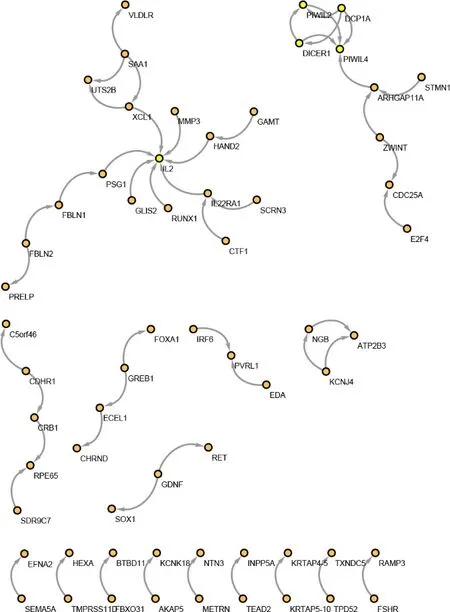
Figure 4 Protein-protein interaction enrichment network

Table 5 Proteins with highest node degrees in PPI analysis
4.DISCUSSION
Liujunzi decoction is widely used to alleviate the side effects of cancer chemoradiotherapy,and has been found to significantly alleviate digestive tract symptoms after chemotherapy in the clinic.8-10In the theory of traditional Chinese medicine (TCM),the efficacy of Liujunzi decoction is believed to beQi-reinforcing in the spleen and stomach,which is usually regarded as improving the function of the digestive tract to enhance resistance to diseases.Studies have shown that it alleviates anorexia11and chemotherapy-induced nausea and vomiting.9,12in cancer.In terms of anti-cancer effects,Liujunzi decoction can inhibit cell migration and invasion,resulting in enhanced epithelial-mesenchymal transition.13The main active ingredients of Liujunzi decoction are baicalein,atractylodes lactone,poria acid,and ginsenoside.Baicalein exerts anticancer effects through a variety of biological pathways.14-16However,the function of a TCM formula is usually not equal to the superposition of single components.Because of the misunderstanding of the differences between the superposition of functions of single components and the function of the whole formula,the therapeutic mechanism of TCM compounds remains unclear.Therefore,bioinformatic analysis is of great significance to understand the mechanism.
Our study showed that IL-2 is a critical protein that interacts with multiple proteins and regulates cytokinecytokine receptor interactions in Liujunzi decoctiontreated NSCLC patients after first-line chemotherapy.IL-2 is produced after the activation of T cells,participating in serial killing and perforin regeneration of tumor cells.IL-2 can promote the proliferation and differentiation of immune lymphocytes,induce the generation of lymphokine-activated killer cells,and enhance the activity of natural killer cells to exert anticancer effects.17It is generally believed that Type 1 T-helper cells (Th1) produce IL-2 and IFN-γ to initiate cellular immunity against tumor cells,and Th2 regulates humoral immunity by secreting IL-4,IL-6,and IL-10.
The disturbance of the Th1 and Th2 balance is considered one manifestation of immune response failure in the tumor microenvironment.18It was found thatRadix platycodonisandGlycyrrhizaeradix et rhizome reduced IL-10 while increased IL-2 and IFN-γ in mice with Lewis lung cancer to exert anti-cancer effect by improving immune function.19Although TCM compounds may target Th1 and Th2 to improve the immune system,there have been no reports of IL-2 improvement in patients with NSCLC after Liujunzi decoction treatment.Our study showed that Liujunzi decoction had an anti-tumor effect on NSCLC patients receiving first-line chemotherapy,which may be related to the interaction of cytokines such as IL-2,revealing the pharmacological mechanism of Liujunzi decoction.
PIWIL4,DICER1,PIWIL2,and DCP1A are the critical genes screened out in this study.GO and KEGG enrichment results showed that PIWIL4,DICER1,and PIWIL2 participate in gene silencing by RNA signaling pathways and are associated with biological processes including regulation of gene expression,RNA-mediated gene silencing,and RNA binding.Studies have shown that their expression levels have prognostic value in a variety of human malignant tumors.20-22Notably,piwi protein can promote cell proliferation,inhibit cell apoptosis,enhance cell migration,and maintain genomic stability in cancer cells.23A previous study showed that PIWIL2 was overexpressed in NSCLC tissue,and there was a significant negative correlation between PIWIL2 and disease-free survival.24Importantly,PIWIL2 and PIWIL4 participate in piRNA-mediated gene regulation.As known,piRNA-induced silencing complexes are produced by binding of piRNAs to piwi proteins,which is a germline-specific member of the Argonaute family acting on genes silenced by small RNAs,as shown in Figure 5.MicroRNAs are regulators of posttranscriptional activity,while piRNAs are considered epigenetic regulators involved in tumor angiogenesis,invasiveness,growth,and metastasis.25Studies showed that piR-651 was overexpressed in NSCLC cell lines,26while piR-55490 inhibited the growth of lung carcinoma.27One theory explained that piwi proteins and piRNAs may contribute to an aberrant “stem-like” state induced by the piwi-piRNA complexes,which leads to genomic silencing following the aberrant methylation of DNA.25Researchers also believe that the piwi protein and piRNA cause epigenetic changes in transposon elements and chromatin structures,because piRNAs can silence transposons through DNA methylation while PIWIL4 can modify chromatin through methylation of p16Ink4a locus,which confirms the positive correlation between PIWIL4 and DNA methylation.28In addition to piRNA,tsRNAs such as ts-101 and ts-53 are also associated with PIWIL2,29indicating that the piwi protein is involved in the regulation of small RNA during cancer development.Our study showed that Liujunzi decoction significantly downregulated PIWIL2 and PIWIL4 in NSCLC patients,indicating that the anticancer effect of Liujunzi decoction might be related to the biological process of piRNA;however,this remains to be clarified by further experiments.
Dicer1 is an endoribonuclease and a member of the RNaseⅢ family that specifically recognizes doublestranded RNA and is critical for the maturation of miRNAs and small interfering RNAs (siRNAs).Because of the importance of miRNA in post-transcriptional gene regulation,various cancer-related investigations have focused on Dicer 1.The basis of RNA interference is the production of 20-31 nucleotide siRNAs.These siRNAs are produced by Dicer protein or by Dicer-independent processes and form complexes with the RNA-induced silencing complex,which carries Argonaute proteins.30Mutation of germline DICER1 leads to various hereditary tumor predisposition syndromes.22A study of the DICER1 pathogenic variation in population databases indicated that despite the rarity of most DICER1 syndrome tumors,the pathogenic DICER1 variation is more common than expected based on the known prevalence of DICER1-related tumors.31,32A study showed that Drosha,Dicer,and Ago2 were expressed in human NSCLC-derived cell lines and NSCLC patient samples,indicating that Dicer participates in the progression of NSCLC to its advanced stage.33In addition,the Dicer enzyme can further process tRNA-derived stress-induced RNAs to tRNA-derived fragments,34indicating that Dicer,like the piwi protein,is involved in the biological processes of a variety of siRNAs.Our study showed that Liujunzi decoction downregulated the expression levels of DICER1 in NSCLC patients,suggesting that DICER1 might be an anticancer target of Liujunzi decoction.
In conclusion,in this study,according to the PPI network analysis,IL2,PIWIL4,DICER1,PIWIL2,and DCP1A were the most noteworthy genes.Our findings suggested that the anticancer mechanism of Liujunzi decoction may be related to the biological processes of siRNAs and piRNAs,especially their roles in gene silencing.These findings provide data and theoretical support for understanding the anticancer mechanism of TCM compounds.
5.ACKNOWLEDGEMENTS
We thank Liwen Bianji,Edanz Editing China(www.liwenbianji.cn/ac) for editing the English text of a draft of this manuscript.
6.RERERENCES
1.Hirsch FR,Scagliotti GV,Mulshine JL,et al.Lung cancer:current therapies and new targeted treatments.Lancet 2017;389:299-311.
2.Ferlay J,Soerjomataram I,Dikshit R,et al.Cancer incidence and mortality worldwide: sources,methods and major patterns in GLOBOCAN 2012.Int J Cancer 2015;136: E359-86.
3.Siegel RL,Miller KD,Jemal A.Cancer statistics,2019.CA Cancer J Clin 2019;69: 7-34.
4.Chen D,Zhao J,Cong W.Chinese herbal medicines facilitate the control of chemotherapy-induced side effects in colorectal cancer:progress and perspective.Front Pharmacol 2018;9: 1442.
5.Sun LL,Lai HZ,Chen ZZ,et al.Modified Liujunzi decoction alleviates chemotherapy-induced anorexia in advanced non-small cell lung cancer: a propensity score matched case-control study.Chin J Integr Med 2020;26: 256-62.
6.Hirose C,Iihara H,Funaguchi N,et al.Prophylactic effect of rikkunshito,an herbal medicine,for chemotherapy-induced nausea in thoracic cancer patients receiving carboplatin-based chemotherapy.Pharmazie 2019;74620-624.
7.Chen YC,Lin AS,Hung YC,et al.Whole genome gene expression changes and hematological effects of rikkunshito in patients with advanced non-small cell lung cancer receiving first line chemotherapy.Exp Ther Med 2017;14: 2040-52.
8.Kang HJ,Jeong MK,Park SJ,et al.Efficacy and safety of Yukgunja-Tang for treating anorexia in patients with cancer: the protocol for a pilot,randomized,controlled trial.Medicine(Baltimore) 2019;98: e16950.
9.Ohnishi S,Watari H,Kanno M,et al.Additive effect of rikkunshito,an herbal medicine,on chemotherapy-induced nausea,vomiting,and anorexia in uterine cervical or corpus cancer patients treated with cisplatin and paclitaxel: results of a randomized phase Ⅱ study (JORTC KMP-02).J Gynecol Oncol 2017;28: e44.
10.Morishige KI.Traditional herbal medicine,Rikkunshito,for chemotherapy-induced nausea and vomiting.J Gynecol Oncol 2017;28: e57.
11.Terawaki K,Kashiwase Y,Sawada Y,et al.Development of ghrelin resistance in a cancer cachexia rat model using human gastric cancer-derived 85As2 cells and the palliative effects of the Kampo medicine rikkunshito on the model.PLoS One 2017;12: e0173113.
12.Tominaga K,Kido T,Ochi M,et al.The traditional Japanese medicine rikkunshito promotes gastric emptyingviathe antagonistic action of the 5-HT(3) receptor pathway in rats.Evid Based Complement Alternat Med 2011;2011: 248481.
13.Kanda R,Miyagawa Y,Wada-Hiraike O,et al.Rikkunshito attenuates induction of epithelial-mesenchymal switchviaactivation of Sirtuin1 in ovarian cancer cells.Endocr J 2020;67:379-86.
14.Kiartivich S,Wei Y,Liu J,et al.Regulation of cytotoxicity and apoptosis-associated pathways contributes to the enhancement of efficacy of cisplatin by baicalein adjuvant in human A549 lung cancer cells.Oncol Lett 2017;13: 2799-804.
15.Deng X,Liu J,Liu L,et al.Drp1-mediated mitochondrial fission contributes to baicalein-induced apoptosis and autophagy in lung cancerviaactivation of AMPK signaling pathway.Int J Biol Sci 2020;16: 1403-16.
16.Cathcart MC,Useckaite Z,Drakeford C,et al.Anti-cancer effects of baicalein in non-small cell lung cancerin-vitroandin-vivo.BMC Cancer 2016;16: 707.
17.Hu CY,Zhang YH,Wang T,et al.Interleukin-2 reverses CD8(+)T cell exhaustion in clinical malignant pleural effusion of lung cancer.Clin Exp Immunol 2016;186: 106-14.
18.Thorsson V,Gibbs DL,Brown SD,et al.The immune landscape of cancer [published correction appears in Immunity 2019 Aug 20;51(2): 411-412].Immunity 2018;48: 812-30.e14.
19.Zhang W,Li M,Du W,et al.Tissue distribution and anti-lung cancer effect of 10-hydroxycamptothecin combined withPlatycodonis RadixandGlycyrrhizae Radix ET Rhizoma.Molecules 2019;24: 2068.
20.Li W,Gao LN,Song PP,et al.Development and validation of a RNA binding protein-associated prognostic model for lung adenocarcinoma.Aging (Albany NY) 2020;12: 3558-73.
21.Navarro A,Tejero R,Viñolas N,et al.The significance of PIWI family expression in human lung embryogenesis and non-small cell lung cancer.Oncotarget 2015;6: 31544-56.
22.Schultz KAP,Stewart DR,Kamihara J,et al.DICER1 tumor predisposition.In: Adam MP,Ardinger HH,Pagon RA,et al.,eds.GeneReviews®.Seattle (WA): University of Washington,Seattle;April 24,2014: 1-34.
23.Tan Y,Liu L,Liao M,et al.Emerging roles for PIWI proteins in cancer.Acta Biochim Biophys Sin 2015;47: 315-24.
24.Qu X,Liu J,Zhong X,Li X,Zhang Q.PIWIL2 promotes progression of non-small cell lung cancer by inducing CDK2 and Cyclin A expression.J Transl Med 2015;13: 301.
25.Maleki Dana P,Mansournia MA,Mirhashemi SM.PIWIinteracting RNAs: new biomarkers for diagnosis and treatment of breast cancer.Cell Biosci 2020;10: 44.
26.Yao J,Wang YW,Fang BB,Zhang SJ,Cheng BL.piR-651 and its function in 95-D lung cancer cells.Biomed Rep 2016;4: 546-50.
27.Peng L,Song L,Liu C,et al.piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling.Tumour Biol 2016;37: 2749-56.
28.Fathizadeh H,Asemi Z.Epigenetic roles of PIWI proteins and piRNAs in lung cancer.Cell Biosci 2019;9: 102.
29.Balatti V,Nigita G,Veneziano D,et al.tsRNA signatures in cancer.Proc Natl Acad Sci U S A 2017;114: 8071-76.
30.Chalbatani GM,Dana H,Memari F,et al.Biological function and molecular mechanism of piRNA in cancer.Pract Lab Med 2018;13: e00113.
31.Kim J,Field A,Schultz KAP,Hill DA,Stewart DR.The prevalence of DICER1 pathogenic variation in population databases.Int J Cancer 2017;141: 2030-6.
32.Stewart DR,Best AF,Williams GM,et al.Neoplasm risk among individuals with a pathogenic germline variant in DICER1.J Clin Oncol 2019;37: 668-76.
33.Prodromaki E,Korpetinou A,Giannopoulou E,et al.Expression of the microRNA regulators Drosha,Dicer and Ago2 in non-small cell lung carcinomas.Cell Oncol (Dordr)2015;38: 307-17.
34.Huang SQ,Sun B,Xiong ZP,et al.The dysregulation of tRNAs and tRNA derivatives in cancer.J Exp Clin Cancer Res 2018;37:101.
猜你喜欢
杂志排行
Journal of Traditional Chinese Medicine的其它文章
- Flavonoids from traditional Chinese herbs for diabetes in rats: a network Meta-analysis
- In-vitro and in-vivo pharmacological screening of Iris albicans
- Detailed approach toward the anti-hyperglycemic potential of Sterculia diversifolia G.Don against alloxan-induced in vivo hyperglycemia model
- Efficacy of Bushen Culuan decoction (补肾促卵方) on ovarian follicle and follicular granulosa cells in mice with premature ovarian insufficiency induced by tripterygium wilfordii polyglycoside
- Efficacy of Wumei Baijiang prescription (乌梅败酱方) on regulatory T cells/ helper T cells Immune balance in mice with ulcerative colitis
- Shenweifang-containing serum inhibits transforming growth factorβ1-induced myofibroblast differentiation in normal rat kidney interstitial fibroblast cells
