Discovery of a wild, genetically pure Chinese giant salamander creates new conservation opportunities
2022-06-07JingChaiChenQiLuMuRongYiNianHuaDaiXiaoDongWengMingXiaoDiYongPengYongTangQingHuaShanKaiWangHuanZhangLiuHaiPengZhaoJieQiongJinRuJunCaoPingLuLaiChunLuoRobertMurphyYaPingZhangJingChe
Jing Chai, Chen-Qi Lu, Mu-Rong Yi, Nian-Hua Dai, Xiao-Dong Weng, Ming-Xiao Di, Yong Peng, Yong Tang,Qing-Hua Shan, Kai Wang, Huan-Zhang Liu, Hai-Peng Zhao, Jie-Qiong Jin, Ru-Jun Cao,0, Ping Lu, Lai-Chun Luo,Robert W.Murphy,8,*, Ya-Ping Zhang,*, Jing Che,9,*
1 State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming,
Yunnan 650223, China
2 Institute of Biological Resources, Jiangxi Academy of Sciences, Nanchang, Jiangxi 330096, China
3 Jiangxi Jiulingshan National Nature Reserve, Jing’an, Jiangxi 330600, China
4 Sam Noble Oklahoma Museum of Natural History & Department of Biology, University of Oklahoma, Norman 73072, USA
5 The Key Laboratory of Biodiversity and Conservation of Aquatic Organisms, Institute of Hydrobiology, Chinese Academy of Sciences,Wuhan, Hubei 430072, China
6 School of Life Science, Henan University, Kaifeng, Henan 475001, China
7 Jing’an County Bureau of Agriculture and Rural Affairs, Jing’an, Jiangxi 330600, China
8 Centre for Biodiversity and Conservation Biology, Royal Ontario Museum, Toronto, Ontario M5S 2C6, Canada
9 Yunnan Key Laboratory of Biodiversity and Ecological Security of Gaoligong Mountain, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China
10 Kunming College of Life Science, University of the Chinese Academy of Sciences, Kunming, Yunnan 650204, China
ABSTRACT Effective conservation of threatened biota relies on accurate assessments and scientific guidance.As an unfortunate example, Chinese giant salamanders(Andrias, CGS) remain critically endangered in nature.Misguided conservation efforts, e.g.,commercial propagation and releasing of millions of likely non-indigenous or interspecific hybrids, have further compromised conservation initiatives.Limited information on wild populations of CGS poses a significant conservation challenge.Following 18-month long field monitoring, we now report the discovery of a wild population of CGS in a closed nature reserve in Jiangxi Province, China.Genomic assessments reveal its genetic distinctiveness and do not detect genetic admixture with other species.Based on morphological and molecular evidences,we describe this CGS as a new species Andrias jiangxiensis sp.nov.This is the only known species of CGS today with a genetically pure, reproducing, in situ population.This discovery emphasizes the important role that closed nature reserves play inprotecting species, and the necessity of integrating long-term field monitoring and genetic assessments.It sets a new pathway for discovering and conserving endangered species, especially for those biotas that are similarly being extirpated by anthropogenic translocations and overexploitation.
Keywords: Conservation; Human translocation;Genetic homogenization; Field monitoring;Taxonomy
lNTRODUCTlON
Effective conservation of threatened biota requires accurate assessments of taxonomy, population distinctiveness, and evolutionary processes, among other data (Crandall et al.,2000; Fraser & Bernatchez, 2001; May, 1990).Such information leads to forming effective management units and conservation decisions (Crandall et al., 2000; Fraser &Bernatchez, 2001).Unfortunately, many critically endangered species are facing extinction in the wild before being assessed.This is especially concerning in the current Anthropocene defaunation that is being driven by habitat destruction, overexploitation, anthropogenic translocation, and climate change (Liu & Weng, 2014; Shaffer et al., 2015; Wang et al., 2021).Very concerning, misinformed conservation initiatives may exacerbate this unfortunate trend (Forcina et al., 2021; Yan et al., 2018).
The mismanagement of Chinese giant salamanders(Andrias, CGS) provides a tragic example of species being extremely endangered in nature, while millions exist in overstocked farms (Cunningham et al., 2016; Lu et al., 2020a;Turvey et al., 2018; Yan et al., 2018).Attaining a body length of almost 2 meters and weighing up to 95kg, the enigmatic CGS are the world’s largest living amphibians (Zhang et al.,2002).Occupying a prominent place in the conservation of global biodiversity, they appear in Appendix I of CITES(CITES, 2021), are listed as Critically Endangered by the IUCN (Liang et al., 2004), and in China have “Class II”protection (National Forestry and Grassland Administration of China, 2021).
Great effort and expense have been spent to save CGS.Nature reserves have been established since 1980s (Ministry of Ecology and Environment of the People's Republic of China, 2017),ex situpropagation has been pushed since the first report of artificial breeding (Yang & Liu, 1978), and population augmentations have been sanctioned since 2002(Shu et al., 2021).These initiatives assumed CGS are one species, but recent studies have revealed that there are from five to eight genetically distinct evolutionary species (Liang et al., 2019; Yan et al., 2018).AlthoughA.davidianus(Blanchard, 1871) andA.sligoi(Bòulenger, 1924) had been described, the limited information of both species could not inform any conservation initiatives.The genetic identity ofA.davidianusremains uncertain because two evolutionary species (clades B and C of Yan et al., 2018) occur proximate to the type locality (Blanchard, 1871), and no wild population has been reported around the regions during the past 20 years.Further complicating efforts, the type locality ofA.sligoiis unknown because the holotype was a translocated animal obtained in the Hong Kong Botanical Gardens (Bòulenger,1924; Turvey et al., 2019).Although the matrilineal identity ofA.sligoiwas assigned to clade D (Turvey et al., 2019; Yan et al., 2018), a handful of wild individuals in its supposed range were verified as translocated species from Shaanxi while the farm-bred ones in Guizhou were genetically polluted quite extensively (Turvey et al., 2019; Yan et al., 2018).Thus, both named species have no genetically verified living population either in the wild or in captivity.
China has established 47 nature reserves since the 1980s to protect CGS and their habitats (Ministry of Ecology and Environment of the People's Republic of China, 2017).However, most reserves have inadequate investigations and assessments of their biota.Field surveys from 2013 to 2016 performed mostly outside of reserves found only 24 individuals of CGS in the wild (Turvey et al., 2018).The surveys covered 97 counties, and averaged 7 person-days per county per year(3.12 person-hours of active searching/county/year, and 6.88 person-days of passive searching/county/year).Unfortunately,these are biased assessments: Guizhou had more than 34%of total surveyed sites, which exceeded efforts in the other 15 provinces (Turvey et al., 2018).Further, no historical records of CGS existed for about one third of the surveyed counties,which were selected by habitat suitability modelling.The unsurprising absence of CGS in these areas magnified rarity.Thus, the extent of local extirpation from nature remains uncertain.
Extremely successfulex situpropagation has occurred since the 2000s almost exclusively in commercial farms(Cunningham et al., 2016).This led to artificial interspecific hybridization via the translocation of non-indigenous salamanders before the existence of multiple species was documented, such as in Guizhou (Yan et al., 2018).Even worse, over 280 000 farm-bred CGS have been released back into the wild without concerns of their genetic identity (Lu et al., 2020a; Shu et al., 2021; Turvey et al., 2018; Yan et al.,2018).Because of potential high profits in farming CGS, illegal activities resulted in a “salamander rush” (Wang et al., 2004;Zhang et al., 2002), analogous to the infamous California Gold Rush of 1848-1855.The existence of five to eight genetically distinct evolutionary species (Liang et al., 2019; Turvey et al.,2019; Yan et al., 2018) and extensive genetic admixture due to anthropogenic translocations (Yan et al., 2018) complicate conservation initiatives.
Past conservation efforts based on the “one species notion”have been ineffective, and both the described and undescribed species of CGS in the wild are suffering from limited information on their genetic identity, population size,distribution, habitat characteristics and other essential aspects of their natural history.Do wild populations free of genetic pollution still exist? To save the species, it is necessary to counter past mistakes, and in doing so, to possibly set a roadmap for saving other endangered species.
Taking new approaches, we focused field surveys in closed nature reserves within the CGS’ historically known distribution,and in areas largely void of commercial CGS farming.Based on Passive Integrated Transponder (PIT) tags and genetic assessments for captured individuals, we discovered a wild,indigenous population of CGS free of genetic pollution in a closed nature reserve in Jiangxi, China.This finding fosters hope for saving CGS and provides the first opportunity for investigating the status of a genetically pure, wild CGS.Herein, we report long-term field monitoring that assesses the natural history, population distinctiveness and morphology of CGS and lay a foundation of effective conservation.
MATERlALS AND METHODS
Field surveys
Long-term field monitoring surveys were conducted from September 2020 to March 2022, in a protected area in Jiulingshan National Nature Reserve, Jing’an County, Jiangxi,China.Our surveys covered periods of peak-activity of juveniles and adults (May-October) (Okada et al., 2008;Turvey et al., 2018), and the initial appearance of newly hatched larvae (Liang et al., 2019; Wang et al., 2017).Captured individuals were PIT tagged and then released at the location of capture.We also conducted field surveys using both active and passive searching methods outside of the protected area but within the historical distribution in Jing’an County, which is densely populated by humans (Figure 1).To avoid poaching and destruction of habitat, we do not provide detailed locality data out of the nature reserve.
Sample collection
Tissue collection mainly involved cutting a small piece of skin from the tail margin.A total of 118 individuals, including 68 juveniles, 40 adults and 10 larvae were sampled in the wild.We also randomly sampled 53 individuals from two farms out of the nature reserve.All sampled tissues were preserved in 95% ethanol and subsequently stored at -80 °C.
For the later morphological comparisons, four specimens were collected from a farm based on the prior genetic assessments (see Methods), and euthanized using benzocaine.These specimens were dissected to determine sex, and liver tissue was taken.The specimens were then fixed with 10% formalin for 72 hours and subsequently transferred to 75% ethanol.Voucher specimens were deposited in the Museum of the Kunming Institute of Zoology(KIZ), Chinese Academy of Sciences (CAS).
Laboratory protocols
Total genomic DNA was isolated from tissue samples using the standard three-step phenol-chloroform extraction method.A mitochondrial DNA (mtDNA) fragment of the gene encoding cytochrome oxidase subunit I (COI) was sequenced for all individuals, and used to assigned individuals to mtDNA matrilines A-E, or U1-U2 (Yan et al., 2018).
As the genome size of CGS is about 50 Gb and a reference genome was not available, we used reduced-representation genome sequencing (RRGS) to obtain genomic sequence data, following the protocols by Yan et al.(2018).After the verification of matriline by mtDNA (see Methods), libraries for 28 individuals were successfully constructed and sequenced,including 18 wild-caught and 10 farm-bred individuals.Data gathering used Specific Locus Amplified Fragment sequencing(SLAF-seq) (Sun et al., 2013).Laboratory work was performed using the protocol described previously (Yan et al., 2018).Reduced complexity libraries were created with genomic DNA using EcoRV-HF restriction enzyme digestions.
Genetic structure analyses
Genetic structure analyses combined SLAF sequences for clades A-E (Yan et al., 2018) and the RRGS data from Jiangxi.All analyses followed the pipeline of Yan et al.(2018).Quality control for raw SLAF sequences involved removing restriction enzyme cutting sites, construction of consensus loci, genotype calling, SNP-filtering, genotype clustering and population structure visualization.
We inferred the phylogenetic tree based on SNPs utilizing concatenation methods, which were performed using maximum likelihood (ML) in RAxML v.8.1.15 (Stamatakis,2014).The ML analyses were executed using 100 rapid bootstrap inferences with a thorough ML search using the GTRGAMMA model.
We used a Principal Coordinate Analysis (PCoA) to visualize population structure based on the genomic SNP dataset using GCTA v1.25.3 (Yang et al., 2011).We inferred individual ancestries of CGS using ADMIXTURE v1.23(Alexander et al., 2009).We sampled one SNP per contig from the original VCF files and filtered this subset by excluding sites with missing genotypes or with a minor allele frequency ≥0.01.ADMIXTURE was run with 10 000 bootstrap resampling at all K values from 1 to 8.The replicate with the highest biological meaning was assumed to have the lowest observed cross validation (CV) error.The PCoA and ADMIXTURE analyses defined six genetically distinct species, which corresponded to clades A, B, C, D, E, and U2.The population from Jing’an County was assigned to clade U2.
Morphological data
Except for one new trait (suborbital distance, SOD),morphological terminologies mainly followed Turvey et al.(2019).Morphological measurements were taken as follows:snout-vent length (SVL), measured from tip of snout to posterior edge of vent; total length (TTL), measured from tip of snout to tip of tail; tail length (TAL), measured from posterior edge of vent to tip of tail; tail height (TAH), greatest height of tail including tail fin; head length (HL), measured from rear of mandible to tip of snout; head width (HW), measured at widest point; greatest diameter of eye (EL); widest diameter of naris(NL); interocular distance (IOD), shortest distance between orbital borders; distance from front of eye to nostril (END);suborbital distance (SOD), measured as vertical distance from inferior edge of eye to inferior edge of upper lip; internarial distance (IND); distance between axillae on left-hand side of specimen (AXD); length of forelimb (FLL), measured from point of limb insertion to tip of longest finger; length of posterior limb (PLL), measured from point of limb insertion to tip of longest toe; first finger length (FI), measured from base of second finger to tip of first finger; second finger length (FII),measured from base of first finger to tip of second finger; third finger length (FIII), measured from base of second finger to tip of third finger; fourth finger length (FIV), measured from base of third finger to tip of fourth finger; first toe length (TI),measured from base of second toe to tip of first toe; secondtoe length (TII), measured from base of first toe to tip of second toe; third toe length (TIII), measured from base of second toe to tip of third toe; fourth toe length (TIV), measured from base of third toe to tip of fourth toe; fifth toe length (TV),measured from base of fourth toe to tip of fifth toe.With the exception of SVL, TTL, TAL, and AXD, which were measured with a tape ruler to the nearest 1 mm, all remaining measurements were taken with a digital caliper to the nearest 0.1 mm.
In addition, morphological data of congeners were obtained from literature, and museum abbreviations included the following: Muséum national d’Histoire naturelle, Paris, France(MNHN); and the Natural History Museum, London, U.K.(BMNH).
lnterviews
We conducted 50 interviews from within 1 km of surveyed rivers in Jing’an County following Tapley et al.(2017).As the selling prices had fallen substantially across China, and the farm-bred offspring of CGS are legal for human consumption in China, CGS is no longer considered a luxury food, and is even available in places such as Walmart.Some interviewed people admitted they had eaten CGS one or two times out of curiosity, but not as a staple.Thus, we added: “do you and your family have the habit of eating CGS” to assess local market demand.We also interviewed farm owners for information on farming.Because some local people stated they could distinguish the indigenous and introduced CGS species by morphological traits, we also asked if the farms had introduced CGS species, and if so, where they came from, and if not, their reasons for breeding indigenous species only.
RESULTS
Discovery of a wild CGS population
According to our 18-month field monitoring, we discovered a native, wild, reproducing population in Jiulingshan National Nature Reserve, Daqi Mountain, Jiangxi, China (Figures 1, 2).Covering 6.50 km of riverine habitat with efforts of 146.40 person-hours, we found 291 juveniles/adults with body lengths of 23-85 cm and masses of 95-4 365 g (Figures 1, 2;Supplementary Table S1).The searching efforts for juveniles and adults represented a catch-per-unit-effort (CPUE) of 0.50 hour/individual (Figure 2A; Supplementary Table S1).An estimation of population size awaits further monitoring and additional surveys outside the current range.Notwithstanding,the discovery of various sized individuals and their rates of recapture (22.52%) inferred a substantial population size in the nature reserve (Supplementary Table S1).Further, the discovery of 524 newly hatched larvae in two continuous years documented reproduction and identified critical breeding caves in need of protection (Supplementary Table S2).The activity period of newborn larvae indicated that their initial feeding occurred from December to February, peaking in late January (Figure 2B, C; Supplementary Table S2).In contrast,CGS surveys out of the reserve in two nearby densely populated towns (Guanzhuang and Zhongyuan) required greater effort (CPUE=17 hours/individual; Figure 1).We didnot observe poaching activities, such as nocturnal fishing,bow-hooks with baits, nets, traps in the streams, or CGS body parts adjacent to streams.Overall, our results indicated a substantial population size, and underscored the effectiveness and necessity of closed national nature reserves for thein situprotection of CGS.

Figure 1 Map of survey localities in Jing’an County, northwest of Jiangxi, China
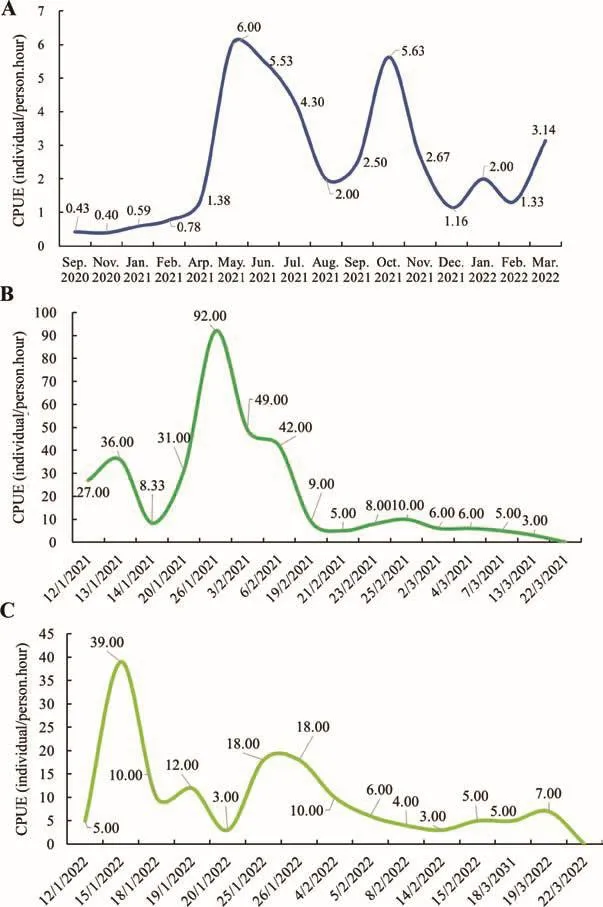
Figure 2 Results of field-monitoring surveys in Daqi Mountain,Jiangxi Jiulingshan National Nature Reserve (Sep.2020-Mar.2022)
Genetic distinctiveness
Genetic assessments suggested that wild-caught CGS preserved their genetic integrity.DNA barcoding of 118 nondestructively sampled individuals revealed that all possessed mtDNA haplotypes of clade U2 (Figure 3A;Supplementary Table S3), indicating no other CGS species had been introduced.These haplotypes were detected previously only in commercial farms (Yan et al., 2018), and they corresponded to clade D of Liang et al., 2019.Clade U2 diverged from its sister clade E from Anhui ca.4.95 million years ago (Ma) (Yan et al., 2018).
As no other evolutionary species had been introduced, 18 wild-caught individuals, including seven adults and 11 juveniles were randomly chosen and sequenced for genomic SNPs.Analyses based on 52 556 SNPs, including ML tree,assessment of genetic structure and PCoA, supported the population’s genetic distinctiveness (Figure 3B-D).The ML tree resolved clades A, B, C, D, E and U2 as distinct groups(Figure 3B), but the PCoA located one individual of clade C from the Pearl River away from two other individuals(Figure 3C, D).Genetic clustering (K=2-8) based on genomic SNPs from six clades by ADMIXTURE analysis indicated seven distinctive groups, in which the best K=7 also supported divergence within clade C.As we only reported the replicate with the highest biological meaning that was consistent with the mtDNA clades, subgroups potentially existing within clades B, C and E (K=6-8) awaited more samples for further determination (Figure 3C-E).Notwithstanding, all K values did not detect genetic mixing of clade U2 and the other species(Figure 3E), indicating they were neither affected by introductions from large-scale commercial farms, nor the hybridization by artificial propagation.
We located two small-scale farms in the towns Guanzhuang and Zhongyuan, which lie within the historical distribution area of CGS and out of the nature reserve.DNA barcoding verified that all 53 individuals in these farms also were from clade U2(Supplementary Table S3).SNP analyses of 10 randomly chosen individuals indicated that they were purebred individuals (Figure 3B-E).

Figure 3 Genetic distinctiveness of CGS from clade U2 found in Jing’an County, Jiangxi, China
The Jiangxi CGS is paraphyletic with respect to the clades A-E, and it differs genetically and morphologically from its Chinese congeners (see below).Thus, herein we describe theJiangxi CGS as a new species.This is now the only species of CGS known to have a genetically pure, reproducing population in the wild.
Taxonomic account
Andrias jiangxiensis sp.nov.Lu, Wang, Chai, Yi, Peng,Murphy, Zhang, and Che (Figure 4; Supplementary Table S4)
Holotype:KIZ 037731, adult male (Figure 4),ex situfrom first generation of offspring of the wild-caught parents, which were collected in Guanzhuang Township, Jing’an County, Jiangxi,China.
Paratypes:KIZ 037728, 037730 (adult females), KIZ 037729(adult male), all with the same collection information as the holotype.
Diagnosis:Andrias jiangxiensissp.nov.can be distinguished from its congeners by a combination of the following characters: (1) head length almost equal to width; (2) head and lower jaw relatively smooth, with small tubercles arranged irregularly; (3) lateral neck fold discontinuous with body fold at forelimb insertion; (4) finger III distinctly longer than finger I;and (5) dorsum red-brown or yellow-brown in life, with large,irregular black patches.
Comparisons:Andrias jiangxiensissp.nov.can be distinguished fromA.japonicus(Temminck, 1836) by having smoother head with indistinct tubercles (vs.dense, distinct,large tubercles).Considering that no population has been genetically identified as both pureA.davidianusandA.sligoi,and that farm-bred individuals exhibit genetic contamination(Yan et al., 2018), to avoid confusion due to translocated animals or hybrids, morphological comparisons are constrained to holotypes only.Specifically,Andrias jiangxiensissp.nov.differs fromA.davidianus(holotype MNHN-RA-0.7613, images posted on: http://coldb.mnhn.fr/catalognumber/mnhn/ra/0.7613) by having a smooth head and lower jaw with indistinct, small, irregularly arranged tubercles(vs.head and lower jaw rough, with distinct, regularly arranged tubercles); fromA.sligoi(holotype BMNH 1945.11.7.1., images in Turvey et al.(2019)) by having a smooth skin on the snout and around eyes (vs.with numerous distinct tubercles), and a distinct finger length formula (finger III distinctly longer than finger I vs.finger III equal to finger I).
Description of holotype:Adult male, body size relatively small, SVL 381 mm, TTL 589 mm; tail muscular, thick at base,gradually flattening posteriorly and distally; dorsal fin well developed, ventral fin much reduced, maximum tail height 76.2 mm; tail short, TAL 208 mm, TAL 54.6% SVL, 35.3%TTL.Head dorsally compressed, wide, HL 102% HW; snout truncated, blunt, widen gradually posteriorly, projecting beyond lower jaw in ventral view; jaw muscular, distinctively widen from snout; jaw muscles continuous with two ovoid,convex temporal muscles dorsally posterior to each eye;nostrils small, NL 1.5 mm, anteriorly and laterally positioned,eye-nare distance 27.6 mm.Eyes small, rounded, EL 4.2 mm,2.8 times NL, protuberant in life, lacking eyelids; dorsolaterally positioned, wide apart, IND 31.6% IOD , EL 44.2% SOD.Mouth large, corner almost reaching mid temporal protuberances; supralabial fold distinct; extending from posterior edge of mouth to axillary.Skin of head and lower jaw mostly smooth with small tubercles scattered dorsolaterally on temporal head and neck in life, all distributed irregularly,gradually increasing in numbers posteriorly; tubercles indistinct after preservation.Transverse gular fold present,without distinct glands.
Body robust, dorsally compressed but less than head,elongated, AXD 34.1% TTL; costal grooves in life, less distinct after preservation; vertebral groove distinct on posterior 3/4 of body; neck fold strongly developed, from corner of mouth to posterosuperior axillary; lateral body folds strongly developed,anterior end inferior and discontinuous from neck fold at limb insertion point, extending to caudal base; single dorsolateral series of large tubercles just above base of lateral fold on each side of vertebrate from neck to caudal base, more distinct in life, barely visible after preservation; remaining body skin mostly smooth in life.
Limbs short, robust, dorsoventrally compressed, with distinct lateral skin folds, more prominent on posterior limbs;hindlimbs relatively longer and stronger than forelimbs, FLL 10% TTL, PLL 12% TTL.Hands and feet dorsally compressed, finger and toe tips enlarged, rounded; finger and toe tips and ventral hands and feet covered with dark carotin layer; four fingers, no webbing, length formula II>III>I>IV,lateral skin fringe distinct on finger IV, continuous with lateral limb fold; five toes, with rudimentary webbing, toe length formula III>IV>II>V>I, toe III, IV, and V with stronger lateral fringes.
Coloration of holotype:In life (Figure 5), dorsum red-brown,with large continuous or discontinuous irregular black patches,gradually decreasing and broken from midbody to head and tail, a few black spots scattered between large patches;ventral surface grayish brown, with black spots scattered along the mandibular margin only; small white spots with tan edge throughout the body; grayish white around eyes, iris gold with black markings.In preservative (Figure 4), bright tints faded, dorsum and abdomen faded to grayish brown and gray,respectively, white spots with tan edge, black patches and spots remain distinct; horny epidermis on tips of fingers and toes as well as palms and soles brown.
Variation:No significant differences in morphology and coloration were found among all adult specimens of the type series, or between genders.However, a distinctive ontogenetic shift existed in coloration.Juveniles less than 30 cm long were mostly spotted (Figure 6A) instead of having larger patches; after exceeding 40 cm, their coloration pattern resembled the type specimens (Figure 6B).

Figure 4 Holotype of Andrias jiangxiensis sp.nov.(KlZ 037731) in preservative

Figure 5 The holotype of Andrias jiangxiensis sp.nov.(KlZ 037731) in life
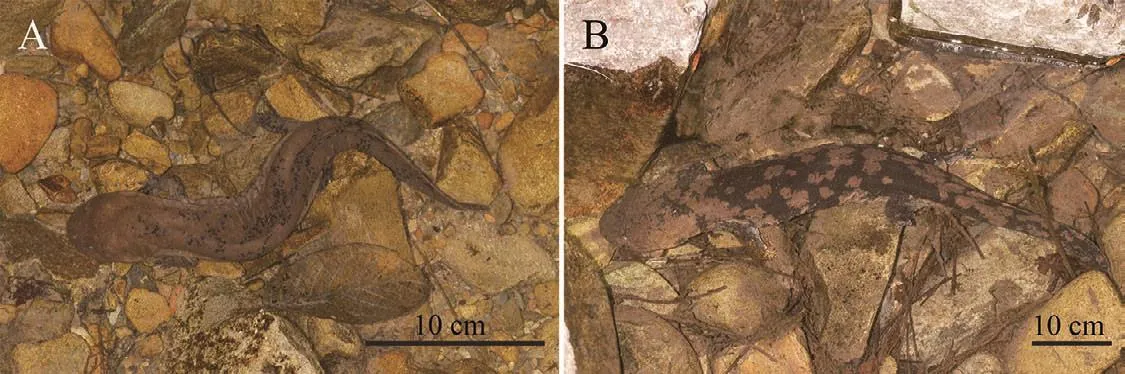
Figure 6 Ontogenetic variation in coloration pattern
Etymology:The specific epithet “jiangxiensis” refers to the type locality of the new species in Jiangxi, China.It denotes the endemicity of the new species to Jiangxi based on our detailed population surveys.We suggest Jiangxi Giant Salamander as its English common name, and 江西大鲵(Pinyin: Jiāng Xī Dà Ní) as its Chinese common name.
Distribution and natural history:Questionnaires with local residents suggest that this species was densely distributed in two towns of Jing’an County before the 1990s.Due to commercial breeding, some indigenous purebred individuals served as breed stock in some local small farms.According to our field monitoring, the wild population of this speciesappears to occur only in Jiulingshan National Nature Reserve.This protected area contains intact habitat, including forested mountains and streams without villages and human activities.It also contains a diversity of teleosts, amphibians, non-avian reptiles, birds and a few mammals.
During the field surveys from September 2020 to March 2022, over 291 wild individuals of adults and juveniles were found in medium-sized, pristine mountain streams with 8-10 m wide, water depth 0.1-1.5 m, and with rocky substrate(Figure 7A, B).Many natural caves formed by fallen wood,tree roots and cracks among rocks on both sides of streams provide hiding places for the new species.As a nocturnal species,A.jiangxiensissp.nov.forages at night.Dorsal color patterning makes it blend well with the rocks at the bottom of the stream, making it difficult to find in flowing water.Vomit and excrement analyses of five wild individuals in July 2021 revealed fish, frogs, aquatic insects, and crustaceans in the natural diet.
Two breeding caves were found in a backwater bay of the stream in January 2021 (Figure 7C, D) and January 2022.A large number of larvae gathered at the entrance, emerging most likely two months after hatching (Figure 7C, D).Larvae sightings peaked in February and exceeded 100 individuals per day.This continued until March, when no more new larvae emerged from the cave, and the larvae outside began to scatter throughout the stream.Combined with a questionnaire to local giant salamander farmers, we speculate that the breeding season ofA.jiangxiensissp.nov.occurs in mid-September.

Figure 7 The habitat of Andrias jiangxiensis sp.nov.in Daqi Mountain, Jing’an County, Jiangxi, China
Conservation:Our long-term field monitoring ofA.jiangxiensissp.nov.yielded only a single incident of a breeding, wild population in the protected area at Daqi Mountain, Jing’an County.This protected area has a riverine habitat of only 6.50 km in linear distance and 36.0 km2of area.The upstream water runs inconstantly throughout the year, as flash floods occur occasionally in summer and droughts often occur in the winter, which lead to frequent habitat fragmentations.The downstream habitat out of the nature reserve is blocked by a dam and is inhabitable toA.jiangxiensissp.nov.due to anthropogenic activities.This small, isolated, and sometimes fragmented population is particularly vulnerable to environmental changes, and thus the species faces a great extinction threat.Therefore, following the IUCN criteria B1 (extent of occurrence less than 100 km2,known to exist at only a single location, and extreme fluctuation of extent of occurrence) (IUCN, 2012), we recommend to classifyA.jiangxiensissp.nov.as Critically Endangered (CR), and we recommend to list the species as Class II protected in China, and to implement further assessments.
Local cultural values:Local cultural pride might have contributed positively to the conservation of indigenous species.Interviews near the survey sites found that most people did not often eat CGS, although nine out of 50 persons had eaten CGS one or two times from curiosity.Six of the 50 interviewed people in Jing’an County, including the towns Guanzhuang and Zhongyuan, could distinguish between the indigenous and introduced CGS species by the shape of head and coloration of adults.They proudly said the indigenous species is “more beautiful” than others (Supplementary Dataset S1); one person described the indigenous species as having a smoother head without tubercles.This differed from negative cultural views of CGS reported in Shaanxi(Cunningham et al., 2016), indicating diverse cultural values across China.
We interviewed the owners of two small-scale farms adjacent to the field survey sites, which have existed for years(Supplementary Dataset S1).They did not cease activities following the dramatic slide in CGS prices from 3 000 RMB/kg(US $470/kg) (Cheng & Zhang, 2015; Shu et al., 2021) to 60 RMB/kg ($9/kg) after 2015.They had refused to use outside sources of CGS (Supplementary Dataset S1), even during the“salamander rush” involving large-scale salamander trading.Such small-scale farms in remote montane villages may be more likely to preserve purebred indigenous lineages than large commercial farms that frequently introduce and sell animals of unknown provenance.They can serve as effective targets for genetic screening, and as reservoirs for potential future population augmentation programs which should such be deemed necessary.
DlSCUSSlON
Our efforts found the only CGS, hitherto known to science, to have a genetically pure, reproducingin situpopulation in China.This discovery is most valuable, especially when the release programs without scientific guidance have causedlarge-scale translocations of CGS (Shu et al., 2021; Turvey et al., 2018; Yan et al., 2018).The surveys in five localities from Anhui, Guangdong, Guizhou, Sichuan, and Zhejiang provinces revealed all captured individuals, including adults and larvae,were translocated animals (Shu et al., 2021; Turvey et al.,2018; Yan et al., 2018).Most of them were verified as released animals and even assigned to three or four different evolutionary species in one locality (Shu et al., 2021).The potential extinction of indigenous species accelerated our actions.The discovery ofA.jiangxiensisgives hope that future investigations will reveal other new species and indigenous populations free of genetic pollution that can be conserved.Assessments of how many other wild populations and habitats exist within the CGS’s historical distribution are urgently needed.However, such small, isolated populations are also more likely to be threatened with extinction (Mace et al.,2008), and protection plans and policies forA.jiangxiensisshould be given priority.This requires greater collaboration between scientists and conservation management departments in both central and local governments.In addition, our interviews revealed the local cultural pride, which might contribute to the conservation of native biotas.This need to be strengthened by developing education and ecological tourism to promote the conservation.
Our analyses document the evolutionary distinctiveness and morphological differences ofA.jiangxiensis.Although artificial interspecific hybridization has been rampant in commercial farms (Yan et al., 2018), genetically distinct species may not choose to hybridize in nature.As with other reports of hybrids having reduced fitness (Cauwelier et al., 2012; Gibeaux et al.,2018; Lu et al., 2020b; Powell et al., 2020), farm-bred hybrids of CGS might be suffering from severe reductions in fitness in F1or F2offspring.The potential genetic incompatibility inex situcrosses of CGS can no longer be neglected, and it is especially critical that F1hybrids not be used henceforth to augment wild populations until the completion of testing for hybrid depression.This also leads us to the urgent need of the broad-scale genetic assessments to identify and PIT tag purebred animals for species-specific augmentation programs.As no genetically pure population ofA.davidianusandA.sligoihas been identified, the new species can only be compared of morphologically with the holotypes of its two Chinese congeners.This problem challenges both scientific research and conservation activities, which need to be overcome in the future.
Our survey methods constitute a new pathway for future investigations.Methodologically, population genomics datasets yield assessments critical for conservation planning and implementation (Crandall et al., 2000; Hohenlohe et al.,2021).Currently, most genetic assessments for CGS were performed by using mtDNA (Liang et al., 2019; Shu et al.,2021), which can only indicate the matrilineal identities rather than the genetic admixture.Thus, it is necessary to employ genomic analyses to screen and identify individuals for conservation; clustering of genomic SNPs can detect extant population structure and identify hybridization (Nater et al.,2017; Schilling et al., 2018).Notwithstanding, population history and the drivers of speciation cannot be inferred without a reference genome and analyses of genomic data.The detections of speciation with gene flow or recent divergence with isolation awaits further investigations based on more modeling and phylogenetic approaches (Martin et al., 2013;Momigliano et al., 2021).
Andrias jiangxiensisprovides the first opportunity to investigate the life history of CGS.In contrast to the comprehensive assessments for Japanese giant salamander and North American hellbender (Hammerson & Phillips, 2004;Kaneko & Matsui, 2004), we had known little about the status of CGS in the wild, especially the habitat ecology and life history, as there is no report of stable wild populations.Fortunately, our efforts forA.jiangxiensisindicated that longterm field monitoring is crucial for documenting the natural history of juveniles, adults, and larvae.These data can reveal the key areas and seasons of feeding and reproduction, and form the guidance of effective conservation actions.
Finally, as opposed to habitats with easy anthropogenic disturbance, our monitoring highlights the roles closed nature reserves in remote montane areas play in future investigations.Our efforts (0.50 hour/individual) were far more successful than the previous surveys outside nature reserve across China (16 weeks/individual) (Turvey et al., 2018), and even more than those for the Japanese giant salamander (A.japonicus, 1.2 hours/individual, including both juveniles and adults) (Okada et al., 2008), and the North American hellbender (Cryptobranchusalleganiensis, 2.2 hours/individual) (Foster et al., 2008).This emphasized the importance of undisturbed habitats for thein situprotection.Efficient metabarcoding in such areas can discover other species also in need of evaluation and protection.Thus, our results form the foundation for evaluating the status of CGS and other biota, especially those similarly suffering from declines caused by anthropogenic translocations and consumption.
NOMENCLATURAL ACTS REGlSTRATlON
The electronic version of this article in portable document format represents a published work according to the International Commission on Zoological Nomenclature (ICZN),and hence the new name contained in the electronic version is effectively published under that Code from the electronic edition alone (see Articles 8.5-8.6 of the Code).This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the ICZN.The ZooBank Life Science Identifiers (LSIDs) and the associated information can be viewed through any standard web browser by appending the LSID to the prefix http://zoobank.org/.
Publication LSID: urn:lsid:zoobank.org:pub:FAAC4B28-F8C8-4ADF-963E-5301A37DEB32
Andrias jiangxiensisLSID: urn:lsid:zoobank.org:act:02CBD 190-DC5E-4087-939C-1BB1A24E8EEF
SClENTlFlC FlELD SURVEY PERMlSSlON lNFORMATlON
Permission for field surveys of CGS in Jiangxi was approved by the Bureau of Fisheries, Department of Agriculture and Rural Affairs of Jiangxi.All fieldwork complied with protocols approved by the relevant forestry offices of Jing’an County,and adhered to all legal requirements.The Kunming Institute of Zoology Animal Care and Ethics Committee approved all protocols of sampling collections and in-person interviews(approval No.IACUC 18 029).
DATA AND CODE AVAlLABlLlTY
Unique haplotypes ofCOIhave been deposited in GenBank under accession Nos.ON150901 and ON150902.Reduced-Representation Genome Sequencing (RRGS) data of the Jiangxi giant salamander have been deposited in the Genome Sequence Archive (GSA) of the National Genomics Data Center (NGDC) at https://ngdc.cncb.ac.cn/gsa/ with accession No.CRA006396.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETlNG lNTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRlBUTlONS
J.Che, Y.P.Z., R.W.M.conceived and designed the research;J.Chai, C.Q.L., M.R.Y., N.H.D., X.D.W., M.X.D., Y.P., Y.T.,Q.H.S., H.Z.L., H.P.Z., J.Q.J., R.J.C., P.L., and L.C.L.conducted field surveys and collected samples; J.Chai,C.Q.L.and M.R.Y.performed molecular work; C.Q.L., K.W.and M.R.Y.measured specimens, and analyzed data; J.Chai and C.Q.L.wrote manuscript, and J.Che, Y.P.Z., R.W.M.discussed and revised the manuscript.All authors read and approved the final version of the manuscript.
ACKNOWLEDGMENTS
We thank Prof.Wen-Xuan Cao for suggestions.Lu-Yang Zhang, Jun Lei, Zheng Li, Ming Jian, and Chuan He helped in the field; Yun-He Wu, Chuan-Xin Yu, Hao-Ping Yin, Shao-Bing Hou, and Wen-Jie Dong helped during lab work.
杂志排行
Zoological Research的其它文章
- Mechanism of hyperproteinemia-induced blood cell homeostasis imbalance in an animal model
- Identification of global alternative splicing and sexspecific splicing via comparative transcriptome analysis of gonads of Chinese tongue sole (Cynoglossus semilaevis)
- Phylogenetic relationships of the zokor genus Eospalax(Mammalia, Rodentia, Spalacidae) inferred from wholegenome analyses, with description of a new species endemic to Hengduan Mountains
- MonkeyTrail: A scalable video-based method for tracking macaque movement trajectory in daily living cages
- Unknown species from China: The case of phrurolithid spiders (Araneae, Phrurolithidae)
- A new species of the gudgeon genus Microphysogobio Mori, 1934 (Cypriniformes: Cyprinidae) from Zhejiang Province, China
