Single-molecule mechanical folding and unfolding kinetics of armless mitochondrial tRNAArg from Romanomermis culicivorax∗
2021-10-28YanHuiLi李彦慧ZhenShengZhong钟振声andJieMa马杰
Yan-Hui Li(李彦慧) Zhen-Sheng Zhong(钟振声) and Jie Ma(马杰)
1School of Physics,Sun Yat-sen University,Guangzhou 510275,China
2State Key Laboratory of Optoelectronic Materials and Technologies,Sun Yat-sen University,Guangzhou 510006,China
Keywords: mitochondrial tRNA, mechanical stability, singlemolecule manipulation, amino acid-chelated Mg2+
1. Introduction
Mitochondria are the sites of aerobic respiration and responsible for energy production in eukaryotic cells. Mitochondrial genomes encode not only proteins essential for energy production, but also parts of the translation machinery,including mitochondrial tRNAs (mt tRNAs).[1]Notably, for bilateral animals, high aerobic respiration demands and accumulative replication errors in asexual reproduction, results in strong mutation pressure in mt DNA encoded genes.[2–6]As a result,mutation in mt tRNAs can cause serious diseases such as myopathies and neurological disorders in human.[7–12]However, inevitable mutations in mt tRNA frequently lead to some bizarre mt tRNAs which have various degrees of truncation and loss on D-or T-arms,being deviation from canonical tRNAs.[13–17]Evidence indicated that these armless tRNAs lacking one or both side arms could still fold into a stable L-shape tertiary structure,[17]and interact with tRNA processing enzymes,i.e.,aminoacyl-tRNA synthetase(aaRS).[18]The conserved tertiary structure is crucial to the functionality of tRNA, including interaction with post-transcriptional editing enzymes, aaRS, and elongation factor, as well as positioning in the ribosome.[19–22]Interestingly, recent studies illustrated an mt tRNA/aaRS recognition mechanism which used shape and folding properties rather than specific base pair in eukaryotic cells to discriminate cognate from non-cognate mt tRNA substrates.[23]Besides the tertiary structure, the mechanical stability of tRNAs is also important to their biological activities. For example,tRNA is under tension and distorted during ribosomal translocation.[24]However,how the armless mt tRNAs maintain the L-shape tertiary structure and mechanical strength is not clear.
A recent research discovered an mt tRNAArgwithout Dand T-arms fromRomanomermis culicivorax, which is the shortest mt tRNA ever known (45 nucleotides (nt), including CCA tail).[15]It has a stem-bulge-stem secondary architecture and an L-shape tertiary structure,but lacks tertiary interaction even in the presence of Mg2+(Fig. 1(a)).[17]Moreover, the predicted folding free energy(−8.34 kcal/mol)for folding mt tRNAArgis substantially higher than canonical tRNA in 1 M NaCl,at 22°C by using MFold.[25]
Besides the advances on structural biology of mt tRNAArgfromRomanomermis culicivorax, its mechanical unfolding/refolding properties have not been studied previously.To clarify its adaptive mechanism of maintaining structural stability in physiological conditions when suffering the environmental destabilization, we performed single-molecule pulling/relaxing experiments on the mt tRNAArgusing homebuilt optical tweezers.[26]Moreover, it is known that RNA is exposed to large amounts of amino acid-chelated magnesium(aaCM)in vivo,[27–30]and these weakly chelated magnesium ions promote the thermal stability of RNA.[31]However,how the aaCM affects the mechanical stability of RNAs is still unknown. Thus, we also investigated the single-molecule mechanical folding/unfolding pathways in the presence of aaCM using optical tweezers. Our results unraveled the folding and unfolding kinetics as well as the free energy landscapes of the mt tRNAArgin different solutions.We discovered the solutiondependent mechanical stability of the bulge region of the armless tRNA,which may shed light on the mechanisms of armless tRNA-protein interactions.
2. Methods and model
2.1. Sample preparation
The synthesis strategy of the single-molecule construct in this study was modified from the one described by Blocket al.[32]In brief, a chemically synthesized DNA containing the mt tRNAArgsequences (42 nt, without 3′CCA tail) and the upstream 1 nt spacer (‘C’) was inserted in between theHindIIIsite andXbaIsite of pUC19 vector(Sangon). The linear DNA template forin vitrotranscription was generated by PCR using the recombinant plasmids, a T7 promoter labeled upstream primer and a downstream primer (see Appendix A,Table A1). RNA containing the mt tRNAArgsequences(42 nt,without 3′CCA tail), the upstream 1 nt spacer sequence, the upstream and downstream 30 nt‘sticky’sequences, was synthesized byin vitrotranscription using T7 RNA polymerase(Promega). Two dsDNA handles were generated by PCR using the pUC19 plasmid as their templates. The 1195 bp upstream handle with an abasic site and a 30 nt 5′overhang was synthesized by PCR, using an autosticky primer and a 5′-digoxygenin modified primer(see Appendix A,Table A1).The 1409 bp downstream handle with a 30 nt 3′overhang was generated by PCR using a 5′phosphorylated primer with three phosphorothioate bonds and a 5′-biotin modified primer (see Appendix A,Table A1),followed by 1 minute lambda exonuclease (New England Biolabs) digestion. All primers were purchased from Sangon, and both handles were purified using PCR purification kit (QIAGEN). The RNA was annealed to the dsDNA handles at the ratio of 1:3:1 in a buffer containing 100 mM NaCl, 20 mM PIPES, and 1 mM EDTA,pH 7.0. During the annealing process, the temperature was first held at 80°C for 5 minutes, then lowered from 80°C to 4°C at a rate of−1°C/min. The samples were first tethered to the cover-glass surface through digoxigenin–antibody interaction and then attached to an 800 nm streptavidin-coated polystyrene bead(Spherotech,Lake Forest,IL,USA)through biotin-streptavidin interaction(Fig.1(b)).
2.2. Amino acid-chelated magnesium(aaCM)
The recipe of aaCM buffer was described by Ryota Yamagamiet al.[31]Briefly,aaCM buffer for 2.0 mM free Mg2+contains 96 mM potassium glutamate, 4.2 mM aspartate, 3.8 mM glutamine, 2.6 mM alanine, 50 mM KCl, 16.0 mM MgCl2, and 20 mM Tris, pH 7.4. All the amino acids were purchased from Sigma-Aldrich.
2.3. Single molecule experiments and data processing
Single-molecule force-ramp experiments were performed using homebuilt single-trap optical tweezers described previously.[26]The 3D piezoelectric stage moved at a constant speed of 100 nm/s during the pulling/relaxing process.The laser power was kept constant during the whole measurement. Each tether was pulled no more than five times. All the experiments were performed at a temperature [(22±1)°C]and humidity[(50±5)%]controlled room. The buffer conditions were 20 mM Tris,0.4 U/µL RNasin plus RNase Inhibitor(Promega),1 mM DTT,in interested KCl and MgCl2concentrations or aaCM,pH 7.4. The 1 kHz raw data were averaged to 200 Hz by using custom MATLAB programs.
2.4. Worm-like chain(WLC)model
During unfolding,contour length changes were found by partitioning the force extension curves(FECs)data into separate states with different contour lengths,then fitting each state to two extensible worm-like chain (eWLC) models in series:one for the dsDNA handles,and the other for the single strand RNA (ssRNA) that is unfolded in each state. We employed a modified Marko–Siggia WLC model described previously as[33]
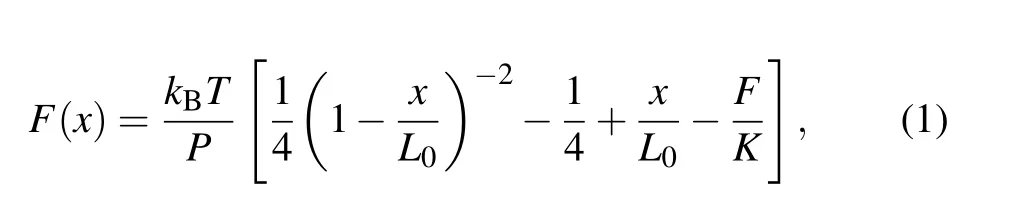
herekBis the Boltzmann constant,Tis the absolute temperature(295 K,22°C),Lis the contour length,Pis the persistence length, andKis the stretch modulus. The parametersL,P, andKdescribing the dsDNA handles were first determined by fitting the FEC for the fully folded state. Then the FECs for the intermediate state and fully unfolded state were fitted by treatingL,P, andKas fixed variables for both the dsDNA handles. The unfolded ssRNALis 0.59 nm/nt,Pis 1 nm andKis 1500 pN,[34,35]respectively. The diameter of an A-form dsRNA helix(2.2 nm).[34]is also taken into consideration when the RNA is fully unfolded.
2.5. Kinetics extracted from force distributions
We assumed the positions of the activation barriers are force-independent,so that the Bell’s kinetic model was applied to describe the force dependence of unfolding and folding kinetics of each transition:[36,37]
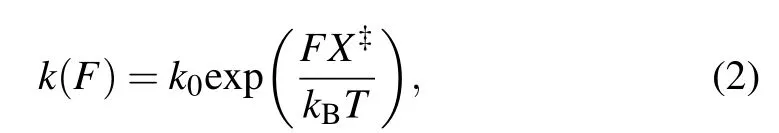
in whichk(F)is the rate constant as a function of forceF,kis the apparent folding/unfolding rate at 0 pN,X‡is the distance from the folded/unfolded state to the transition state along the reaction coordinate,kBis the Boltzmann constant, andTis absolute temperature(295 K,22°C).
The folding/unfolding kinetics can be expressed in a transformed equation by taking the logarithm of Eq.(2):
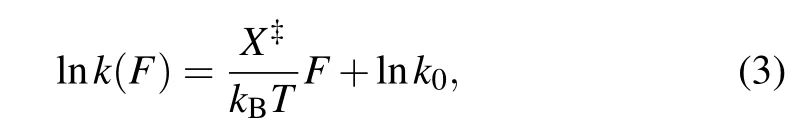
where the slope isX‡/kBT, and theyintercept is lnk. The critical forceF1/2is defined as the force at which the unfolding rate equals to the refolding rate,obtaining from the crossing point of the force-dependent unfolding and refolding rate curves,i.e.,tok1/2.
We determined the force-dependent unfolding and refolding kinetics extracted from distributions of the rupture forces using Dudko’s method.[38]The rate constant at forceF,k(F),can be computed from force probability density histogram:
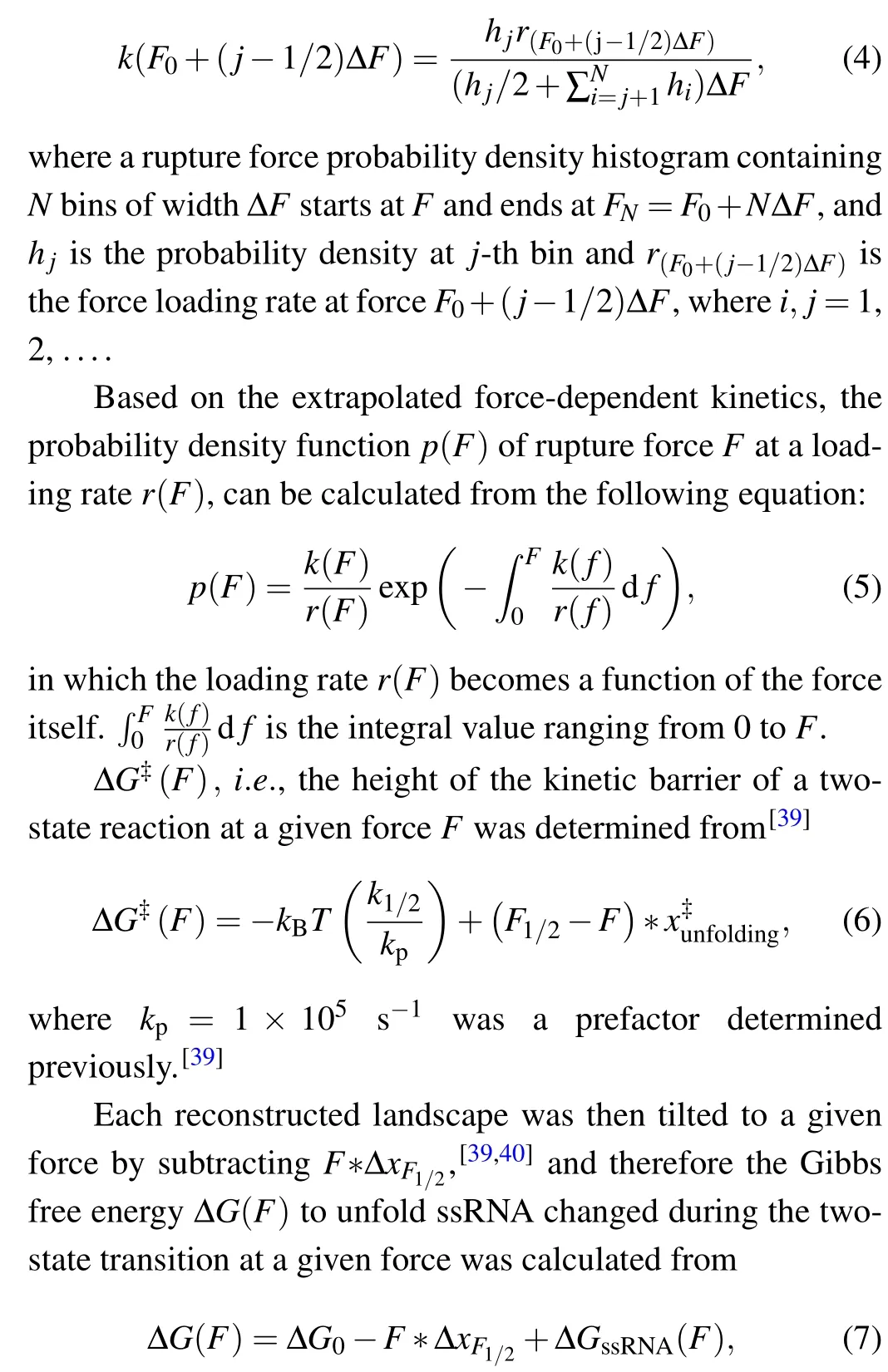
where ∆GssRNA(F) is the free energy of stretching the unfolded ssRNA from 0 pN to forceF, and the free energy at 0 pN ∆G0is determined from

whereW(F)is the reversible work of the reaction by integrating the FEC from the folded state at 0 pN to the unfolded state at forceF, and ∆Gstreching(F) is the free energy of stretching the handles and the unfolded ssRNA from 0 pN to forceF.
3. Results and discussion
We firstly performed pulling/relaxing experiments at 146 mM KCl whose monovalent cation concentration corresponds to physiological conditions. However,we only observed a discernible transition from these trajectories at around 12 pN–14 pN and a suspected transition at about 4 pN, which was difficult to distinguish (Fig. 2(a)). In consideration of the effect of monovalent cation concentration on structural stability,then we increased salt concentration to 1 M KCl and observed two obvious two reversible transitions: a large hopping transition at 7 pN–9 pN with 8 nm–10 nm end-to-end extension change(∆x), and a small back-and-forth transition at 12 pN–14 pN with 4 nm–5 nm ∆x(Fig. 1(c)). It indicated that only an intermediate state(‘I’)was observed between a fully folded state (‘F’) and a fully unfolded state (‘U’). Scarcely hysteresis was observed between pulling and relaxing traces,indicating that the mechanical pulling pathway is highly reversible.We fitted the state ‘F’ by applying Eq. (1) (Fig. 1(c), green curve),whose fitting parameter describes the stretching of the handles. Considering it hard to distinguish the state ‘I’ and state ‘U’ by eWLC fitting, we then employed the theoretical ssRNA length changed of the state‘I’(26 nt)and state‘U’(41 nt) compared to state ‘F’, as well as the fitting parameters of‘F’ states to draw the theoretical pulling curves of these two states (Fig. 1(c), orange and sky-blue curves). These curves are well superimposed onto FECs data,which indicate that the acceptor stem and the bulge are disrupted by tension firstly and anti-codon hairpin as followed(Fig.1(d)).
As can be seen, K+concentration mainly affected the first transition but almost not affected the second transition.As the backbone is negative charged, RNA depends critically on cation ionic conditions which can stabilize RNA secondary and tertiary structures.[41,42]On the basis of 1 M KCl concentration, the addition of 5 mM MgCl2did not apparently affect the mechanical folding/unfolding of mt tRNAArg(Fig. 2(c)). These mechanical unfolding/refolding results agreed with NMR signals measured by Tina J¨uhlinget al.,[17]indicating that the presence of magnesium ion did not induce additional tertiary interactions for mt tRNAArg.
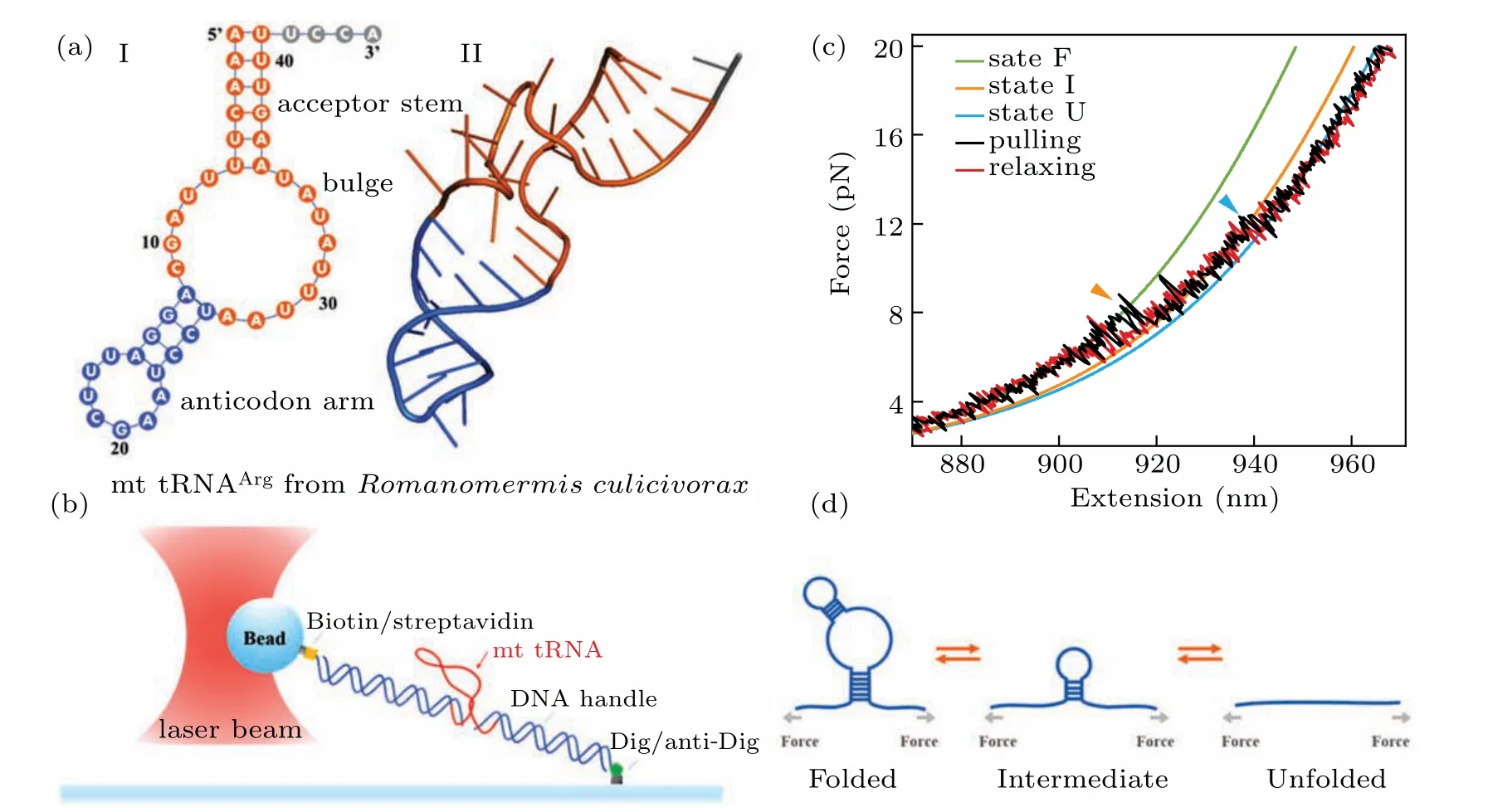
Fig.1. Scheme of the mt tRNAArg mechanical unfolding/refolding experiments.(a)The proposed secondary structure(I)and tertiary structure(II)of mt tRNAArg from Romanomermis culicivorax,which lacks both D-and T-arms.[17](b)The schematic plot of the pulling experiment:mt tRNAArg molecule with two functionalized dsDNA handles was attached between the anti-digoxigenin antibody coated cover-glass surface and a 800 nm streptavidincoated polystyrene bead. (c)Representative force-extension curves(FECs)of unfolding(black)and refolding(red)of mt tRNAArg during the pulling experiments at 1 M KCl. The curves are averaged to 200 Hz from 1 kHz raw data. WLC fitting was applied to the FECs, discovering three states:‘F’,the fully folded state(green); ‘I’,the intermediate state(orange); ‘U’,the fully unfolded state(sky-blue). (d)The probable two-step pathways of unfolding/refolding of the mt tRNAArg.
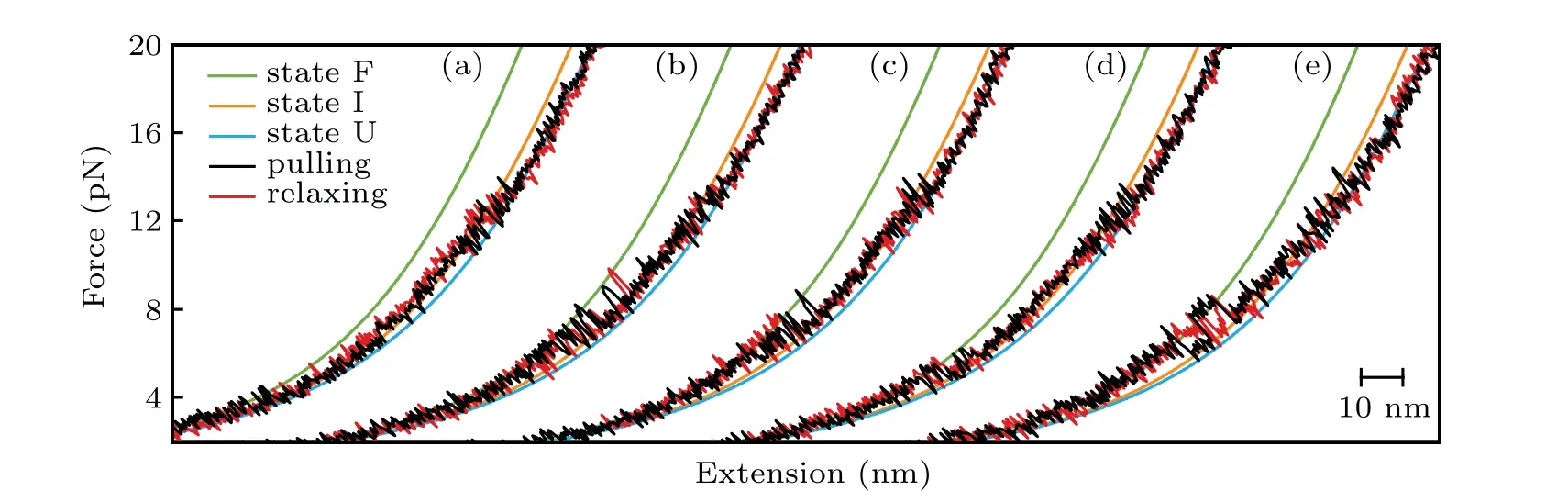
Fig.2. Typical FECs of the mt tRNAArg at different solutions: (a)146 mM KCl; (b)1 M KCl; (c)1 M KCl and 5 mM MgCl2; (d)146 mM KCl and 2 mM MgCl2;(e)aaCM solution including 146 mM K+ and 16 mM total Mg2+ (with 2 mM free Mg2+).
The acceptor stem and bulge structure were stable under high cation concentration instead of physiological concentration,but in fact high cation concentration did not exist in normal cells.To clarify the possibility of mt tRNAArgmaintaining stable structurein vivo,we measured its mechanical unfolding and refolding under a near cellular condition,i.e.,in an amino acid-chelated magnesium buffer(aaCM),which contains 146 mM K+and 16 mM total Mg2+(with 2 mM free Mg2+)(see methods for details).[31]As can be seen,the unfolding rupture forces of the first transition were around 7 pN–9 pN,which is close to those at 1 M KCl (Fig. 2(e)) but significantly higher than those at 146 mM KCl, while aaCM did not obviously affect the second transition. We also performed the pulling experiment at 146 mM KCl and 2 mM MgCl2as a control(Fig. 2(d)). In this case, the unfolding rupture forces of the first transition decreased to 5 pN–7 pN(Fig.2(d)),while those of the second transition did not obviously changed. We also performed the Kolmogorov–Smirnov test(KS test)atα=0.05 level on the unfolding and refolding rupture force distributions of four different solution conditions.We compared the rupture force distributions of two selected solutions in each test. The results indicated that, when the solution condition changes,folding and unfolding rupture forces of F–I but not I–U transition are statistically obvious different(see Appendix A,Table A2).
In addition, we also measured the extension changes at rupture force (∆x) for each unfolding transition (Fig. 3). The measured ∆xvalues were not apparently affected by solution conditions,and they are all well superimposed on the predicted WLC prediction curves for the stretching of unfolded ssRNAs during the transition,which were calculated by Eq.(1). These results suggested that both high concentration of cations and weakly chelated magnesium ions in aaCM (~16 mM total Mg2+and 2 mM free Mg2+) could promote the mechanical stability of armless mt tRNAArgwithout changing the intermediate structure,which suggested that they enhanced the stability of the bulge region from being destructed at lower external forces.
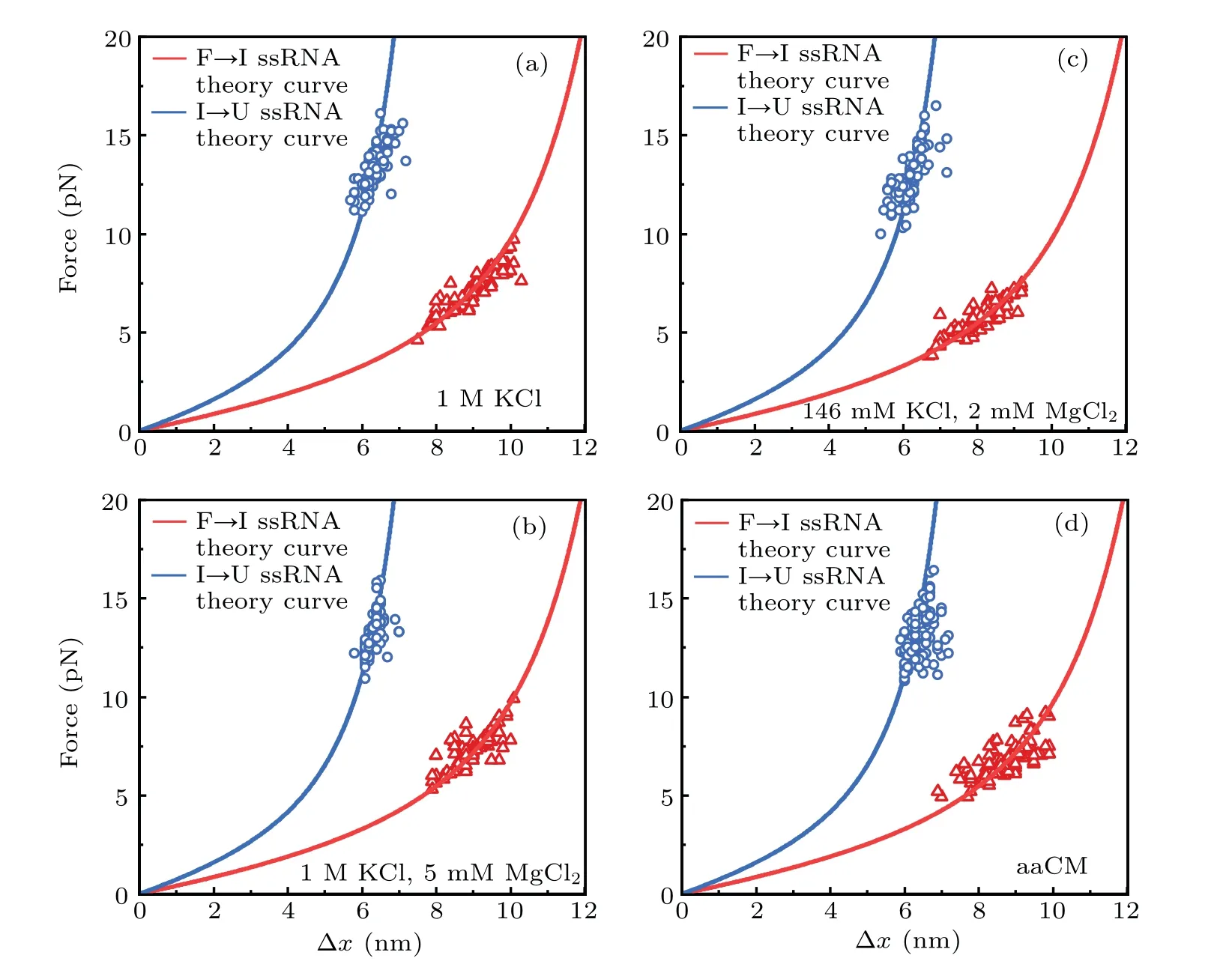
Fig.3. Force-dependent extension changes(∆x)of two unfolding transitions: state‘F’to state‘I’and state‘I’to state‘U’.∆x values of F→I transition(red triangle)and I→U transition(blue circle,2.2 nm was added)are plotted at their rupture forces respectively. Red curves(acceptor stem and bulge,26 nt) and blue curves (anticodon arm, 15 nt) are the ssRNA WLC predictions. Measured data are well superimposed on the WLC predictions in different solutions,including: (a)1 M KCl;(b)1 M KCl and 5 mM MgCl2;(c)146 mM KCl and 2 mM MgCl2;(d)aaCM.
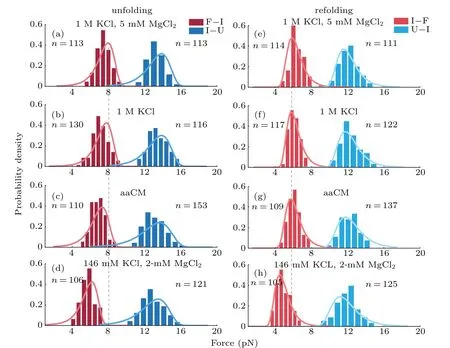
Fig.4. Probability density distributions of unfolding and refolding rupture forces in different buffers. (a)–(d)Unfolding forces in F→I(crimson)and I→U(light-red)transitions. (e)–(h)Refolding forces in I→F(dark-blue)and U→I(sky-blue)transitions. The solid curves were plotted using Eq.(5)and extrapolated kinetics parameters from Table 1 respectively. n is the number of observed transitions.
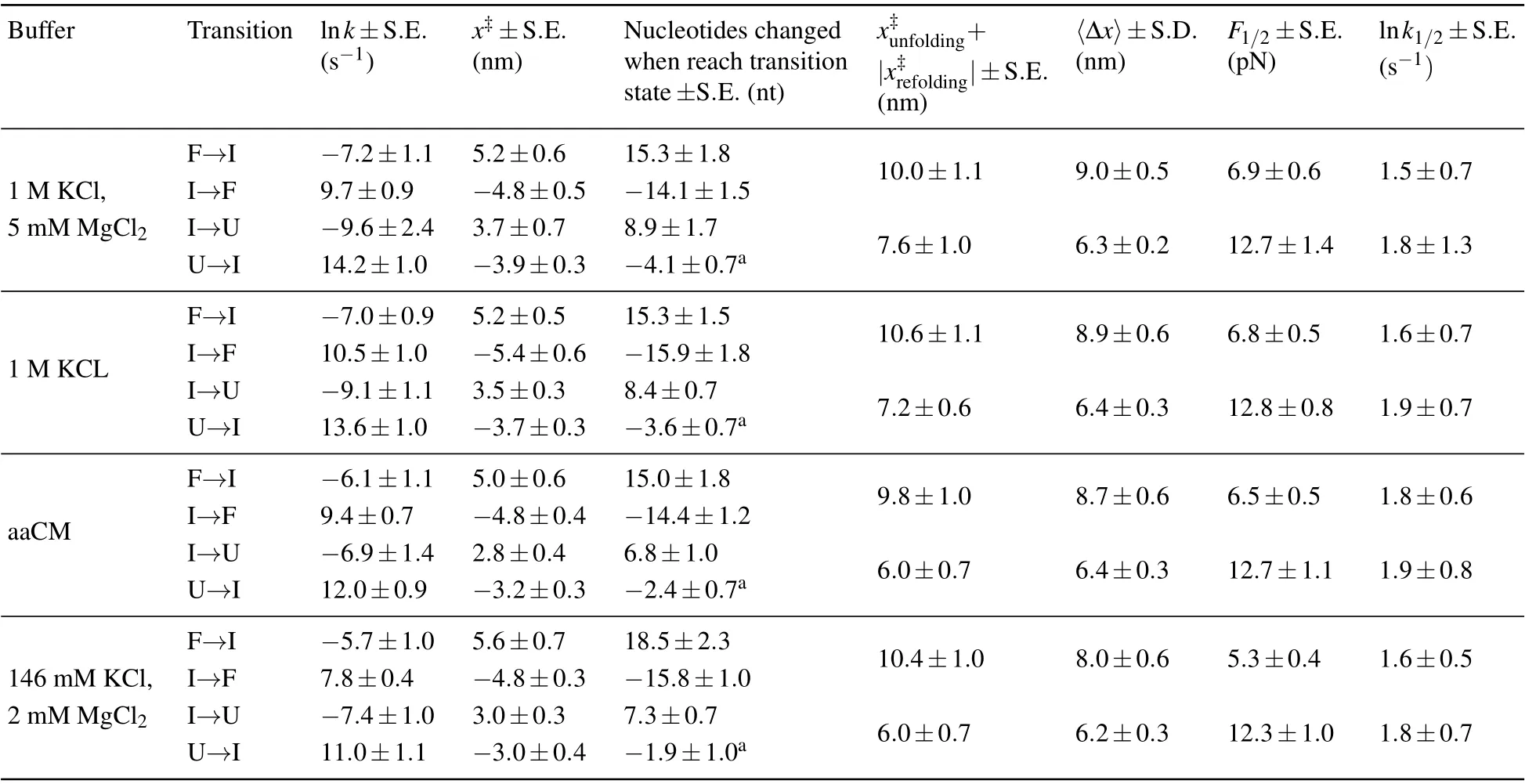
Table 1. Unfolding and refolding kinetics parameters of mt tRNAArg extracted from the force distributions of different solutions. The presented data are mainly from linear fitting by using Eq.(3).
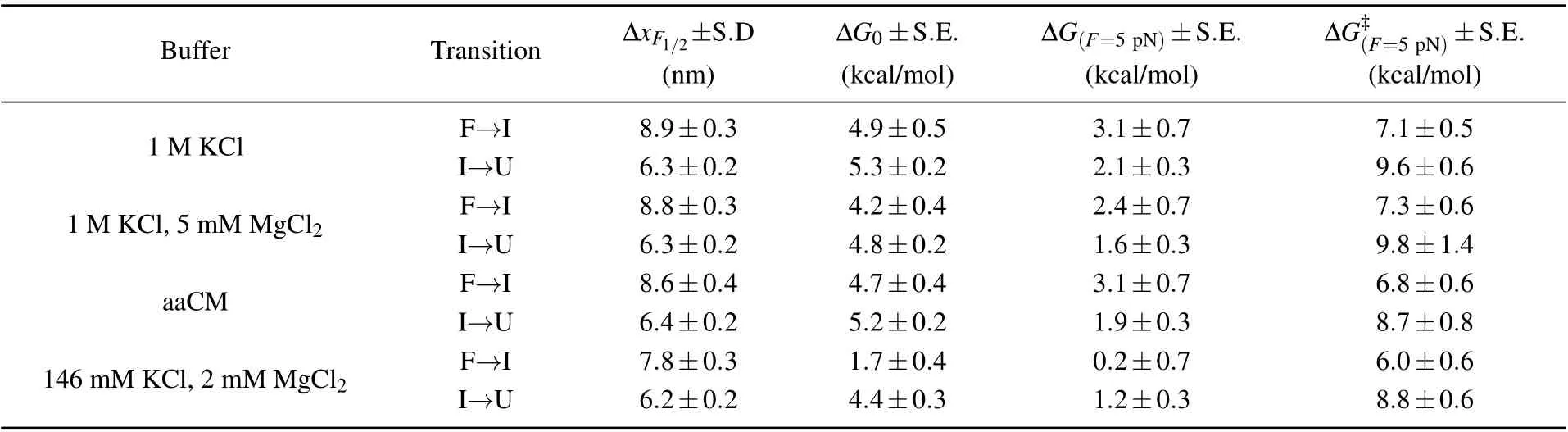
Table 2. End-to-end extension changes at critical force and free energy changes as well as activation energies calculated from parameters shown above in Table 1.
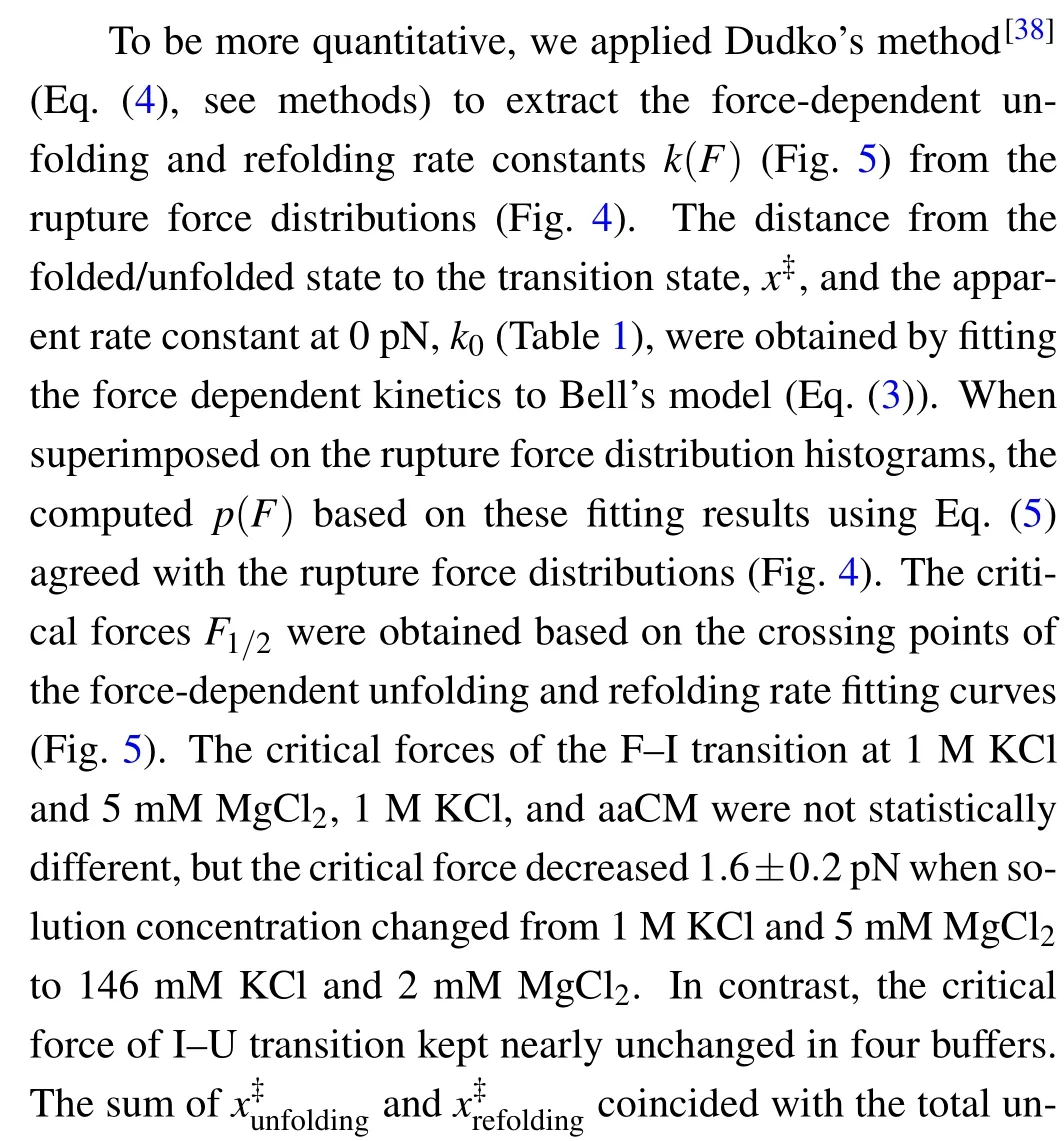
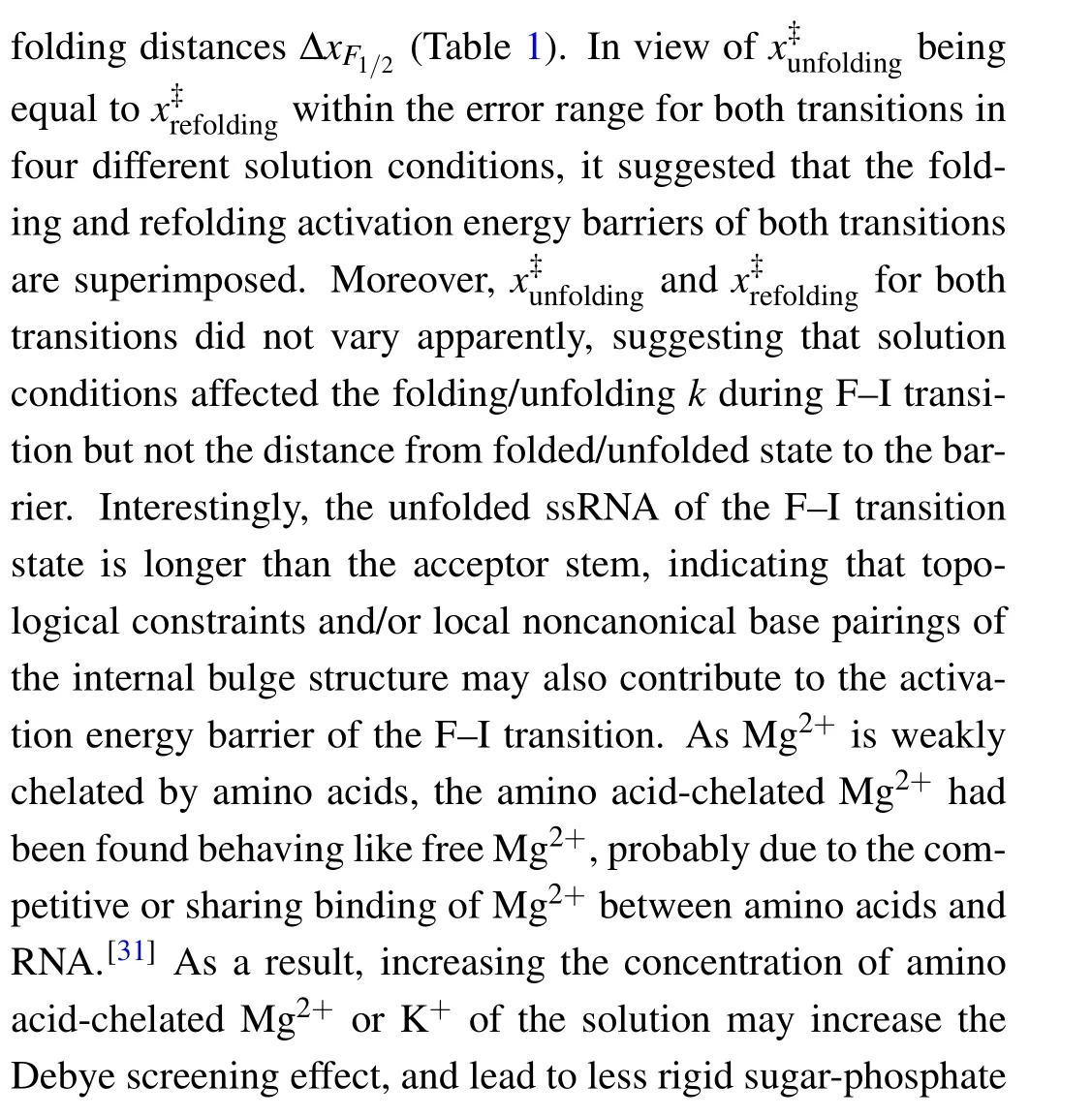
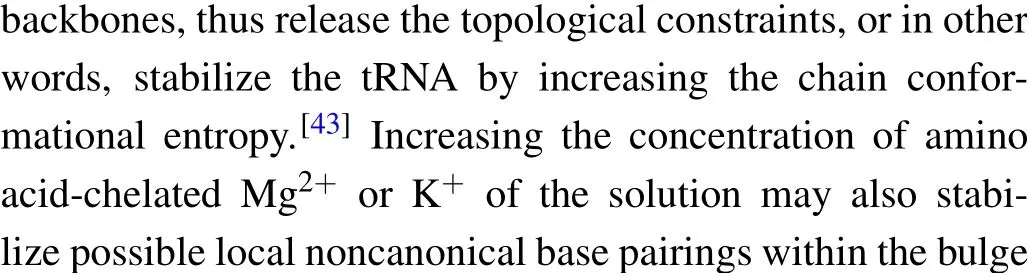

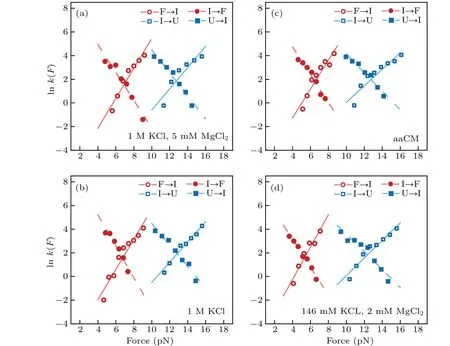
Fig.5. Theforce-dependent unfolding(open markers)andrefolding(fliled markers)kinetics indifferent solution conditions.The criticalforces(F1/2)of two transitions wereobtainedon thecrossing pointsofthe unfoldingandrefolding rates curves. The R2values ofthelinearftis rangefrom 0.87to 0.97.
Moreover, we also reconstructed the three-state free energy landscapes for mt tRNAArgin four solutions. The change of Gibbs free energies ∆G, the height of barrier ∆G‡and the extension of each transition were plotted with reference to the fully unfolded state (state ‘U’) by piecewise two-state analyses of each transition at 5 pN (calculated by Eqs. (6)–(7)).Clearly, the free energy landscapes of the tRNA at 1 M KCl,1 M KCl,and 5 mM MgCl2or in aaCM buffer were not obviously different, while the free energy of state ‘F’ at 146 mM KCl and 2 mM MgCl2was around 2.9 kcal/mol higher than the one at 1 M KCl at 5 pN. In addition, the sum of ∆G0of both transitions (10.2±0.7 kcal/mol) at 1 M KCl is larger than the predicted free energy (8.34 kcal/mol) for unfolding the secondary structures,at 22°C by using MFold,[25]further indicating the existence of possible local noncanonical base pairings within the bulge region. Although the unfolding free energy is higher than canonical tRNA,considerable stability of these mt tRNAArgmolecules has been observed during pulling experiments. Our studies clearly illustrated the presence of aaCM or high concentration of cations could increase the mechanical stability of mt tRNAArgby stabilizing the fully folded state,which further supports the suggestion that aaCM or high concentration of cations increase the mechanical stability of the armless tRNA by stabilizing the bulge region.
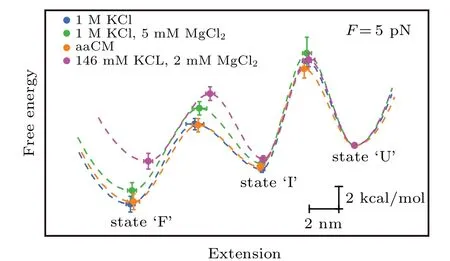
Fig.6. Free energy landscapes for mt tRNAArg. The key features of the energy landscapes for the three-state native unfolding/folding pathways were reconstructed from piecewise two-state analyses of each transition at 5 pN.Energies and positions are plotted with reference to the fully unfolded state(state ‘U’). Error bars show S.E. Dotted lines indicate notional landscape shapes. The free energy of state‘F’at 146 mM KCl and 2 mM MgCl2 was around 2.9 kcal/mol higher than the one at 1 M KCl at 5 pN.
4. Conclusion
In this study, we employed single-trap optical tweezers to perform single-molecule mechanical folding and unfolding experiments on an armless mt tRNAArgmolecules in different solution conditions. We discovered that the armless tRNA followed a highly reversible two-step folding/unfolding pathway with one intermediate in all four different solutions. High concentrations of cations or aaCM can promote the mechanical stability of the armless tRNA, probably by stabilizing the bulge region of the tRNA.
Our studies suggest that the bulge region of the armless tRNA is sensitive to the surrounding electrostatic environment,which could be disrupted by changing the concentration or types of ions in the solution,as described in this study. Moreover,it could also be disrupted by possible post-transcriptional nucleoside modification.[48]As the bulge region functions as a hinge between the acceptor arm and the anti-codon arm,such disruption may change the distance between the aminoacylation site and the anticodon, which is critical to the biological functions of tRNAs.[15]Overall, our studies indicate the critical role of the bulge region in the mechanical stability of the armless tRNAs.
Appendix A:Supplementary information
Some experiment results and tables for better understanding the present article are given below.
We employed a nonparametric test,i.e., Kolmogorov–Smirnov test(K–S test)atα=0.05 significance level for the unfolding and refolding force distributions in four different solutions to ask whether a significant difference between the rupture force distributions of two selected solutions in each test.Here,the rupture forces from two selected solutions were the samples for comparison and we firstly assumed the two samples had no significant difference initially. If the test results rejected the initial assumption atα=0.05 level, the parameterhshould be equal to 1, otherwiseh=0. The results were shown in Table A2.

Table A1. Sequences of oligomers used in the experiments. Here, ‘-’ represents an abasic site, ‘p’ represents phosphorylation and‘*’is phosphorothioate bond.
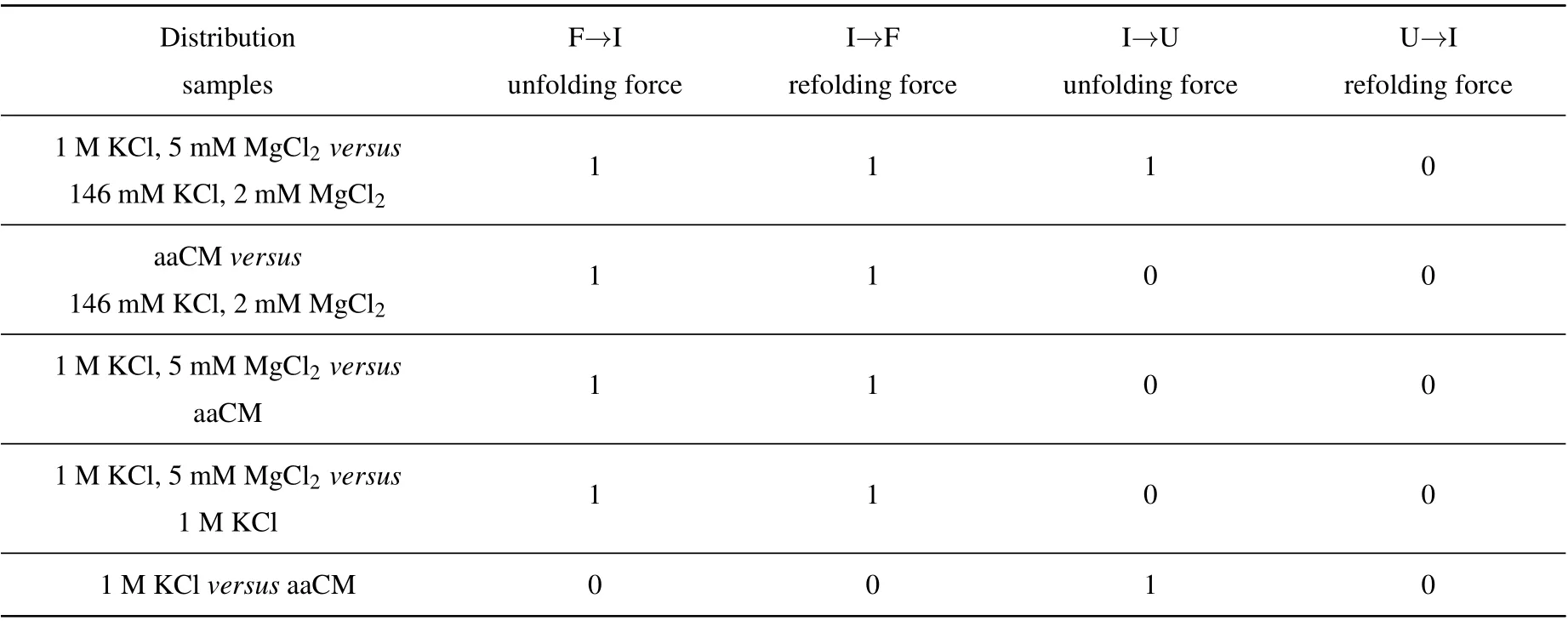
Table A2. The calculated results of parameter h were shown,parameter h=1 consists with significant difference,while h=0 agrees with no difference.
Acknowledgements
We thank members of the J. M. Laboratory for helpful discussions. We also wish to thank for the support from the Physical Research Platform in School of Physics,Sun Yat-sen University(PRPSP,SYSU).
杂志排行
Chinese Physics B的其它文章
- Physical properties of relativistic electron beam during long-range propagation in space plasma environment∗
- High winding number of topological phase in non-unitary periodic quantum walk∗
- Widely tunable single-photon source with high spectral-purity from telecom wavelength to mid-infrared wavelength based on MgO:PPLN∗
- Control of firing activities in thermosensitive neuron by activating excitatory autapse∗
- Adaptive synchronization of chaotic systems with less measurement and actuation∗
- Dynamics analysis of a 5-dimensional hyperchaotic system with conservative flows under perturbation∗
