Diagnostic description and geographic distribution of four new cryptic species of the blue-spotted maskray species complex (Myliobatoidei: Dasyatidae;Neotrygonspp.) based on DNA sequences*
2018-07-11PhilippeBORSAIrmaARLYZAThierryHOAREAUKangNingSHEN
Philippe BORSA Irma S. ARLYZA Thierry B. HOAREAU Kang-Ning SHEN
1Institut de Recherche pour le Développement(IRD),UMR 250“Ecologie Marine Tropicale des Océans Paci fi que et Indien”,BP A5,98848 Nouméa,New Caledonia
2Lembaga Ilmu Pengetahuan Indonesia(LIPI),Pusat Penelitian Oseanogra fi(P2O),Jakarta,Indonesia
3Molecular Ecology and Evolution Programme,Department of Genetics,University of Pretoria,Pretoria,South Africa
4Aquatic Technology Laboratories,Agricultural Technology Research Institute,Taiwan,China
AbstractNine morphologically similar but genetically distinct lineages in the blue-spotted maskray species complex, previouslyNeotrygon kuhlii(Müller and Henle) qualify as cryptic species. Four of these lineages have been previously described asNeotrygon australiaeLast, White and Séret,Neotrygon caeruleopunctataLast, White and Séret,Neotrygon orientaleLast, White and Séret, andNeotrygon varidens(Garman), but the morphological characters used in the descriptions offered poor diagnoses and their geographic distributions were not delineated precisely. The objective of the present work is to complete the description of the cryptic species in the complex. Here, an additional four lineages are described as new species on the basis of their mitochondrial DNA sequences:Neotrygon bobwardi, whose distribution extends from the northern tip of Aceh to the western coast of Sumatera;Neotrygon malaccensis, sampled from the eastern part of the Andaman Sea and from the Malacca Strait;Neotrygon moluccensis, from the eastern half of the Banda Sea; andNeotrygon westpapuensisfrom the central portion of northern West Papua. The geographic distributions ofN.australiae,N.coeruleopunctata,N.orientale, andN.varidensare updated. For each species, a diagnosis is provided in the form of a combination of private or partly-private nucleotides at 2-4 nucleotide sites along a 519-base pair fragment of theCO1gene. We believe that the present taxonomic revision will provide information relevant to the sound management and conservation of cryptic species of the blue-spotted maskray in the Coral Triangle region.
Keyword:molecular taxonomy; diagnosis; distribution
1 INTRODUCTION
Species usually are the fundamental units in biogeography, community ecology and conservation ecology. Since pioneer allozyme studies about five decades ago (Manwell and Baker, 1963), molecular genetic markers have been essential in uncovering cryptic species within nominal species once believed to be widely distributed (Bickford et al., 2007). Cryptic species are a common occurrence in marine ecosystems(Knowlton, 1993; Zemlak et al., 2009; Nygren, 2014;Durand and Borsa, 2015). Ignoring them may have adverse consequences in marine fi sheries management,biodiversity management, and conservation(Krishnamurthy and Francis, 2012; Van Campenhout et al., 2014; Borsa et al., 2015; Pante et al., 2015). It is therefore important to accurately delineate cryptic species. The step beyond, naming species, is essential in biodiversity studies because formally designating a species by its binominal name allows to link and compare the results of scientists working separately on conSpecific samples (Pante et al., 2015). DNA sequences potentially provide a profusion of diagnostic characters, hence are useful tools in modern taxonomy and essential ones to delineate and describe cryptic species (Tautz et al., 2003; Vogler and Monaghan,2006; Cook et al., 2010; Jörger and Schrödl, 2013). In contrast, morphometrics as presented in some recent taxonomic work in Elasmobranchs (e.g., Last et al.,2016) did not offer clear diagnoses. In our view,morphometric descriptions in this case should be undertaken a posteriori, i.e, once species of a complex of cryptic species have been delineated and described based on molecular genetics.
Cryptic species have been reported in morphologically intractable species complexes ofindo-West Pacific fishes (e.g., Zemlak et al., 2009;Durand and Borsa, 2015; Randall and Victor, 2015)including stingrays (Naylor et al., 2012; Arlyza et al.,2013b; Borsa et al., 2013c). In a recent paper (Borsa et al., 2016b), the maximum-likelihood phylogeny ofCO1- andcytochromebgene haplotypes determined nine main lineages within the blue-spotted maskray,previouslyNeotrygonkuhlii(Müller and Henle,1841). Analysis of coalescence patterns (Pons et al.,2006) generally confi rmed that these lineages represent separate species. Also, the lineages are geographically distributed in a parapatric fashion,indicating reproductive isolation (Bull, 1991). Eight of these lineages occur in the Coral-Triangle region and the ninth one occurs in the Indian Ocean, from India to Tanzania (Borsa et al., 2016b). A specimen from Vanikoro in the Santa Cruz archipelago,northeastern Coral Sea has been designated as lectotype ofN.kuhlii(Last et al., 2016) and three of the lineages, all from the Coral Triangle, have been described as new species, namelyN.australiaeLast et al., 2016,N.caeruleopunctataLast et al., 2016 andN.orientaleLast et al., 2016. A fourth one,N.varidens(Garman, 1885) has been resurrected from synonymy withN.kuhlii(Last et al., 2016). None of the nine lineages uncovered by Borsa et al. (2016b) wasN.kuhliiaccording to its recent re-description (Borsa and Béarez, 2016). Five of these nine lineages remain undescribed, illustrating the limitations of morphological characters to distinguish them. Two additional lineages up to now represented each by a single haplotype also require attention (Borsa et al.,2016b).
The present paper is a follow-up of Borsa et al.(2016b), whose purpose was to delineate the cryptic species in the blue-spotted maskray species complex.We emphasize the distinction between species delineation, species description, and species identifi cation, although DNA markers may be used in all three cases (Vogler and Monaghan, 2006; Cook et al., 2010). In the present paper, the nucleotide sequence of a portion of theCO1gene is used to diagnose species in a formal taxonomic description.The same genetic marker is the standard in DNA-barcoding, where it is used as a means ofidentifying individuals to species by sequence similarity (Vogler and Monaghan, 2006; Ratnasingham and Hebert,2007). The aims of the present paper are (1) to describe as new species, on the basis of their DNA sequences, the still-anonymous lineages of the bluespotted maskray previously underN.kuhlii; (2) to provide a molecular diagnosis for each of the nine lineages previously underN.kuhlii; (3) to provide or update their distributions, based on the available nucleotide-sequence data at theCO1- andcytochrome bgene loci.
2 METHOD
To evaluate the diagnosticity of the morphological characters used in the description or re-description ofN.australiae,N.caerulaeopunctata,N.kuhliiandN.orientale(Last et al., 2016), we tabulated their ranges of values in each of the four species. We then assessed whether a given character enabled the distinction of at least one of the species from the other species by looking at possible disjunctions in the ranges of values.
All individuals included in the present survey along with sampling details, accession numbers in the GenBank nucleotide database (http://www.ncbi.nlm.nih.gov/) and registration numbers in the Barcoding of Life Datasystem (BOLD; http://www.barcodinglife.com/; Ratnasingham and Hebert, 2007) are listed in Borsa et al. (2016b). Voucher specimens of bluespotted maskray were deposited at the Museum Zoologicum Bogoriense (MZB) in Cibinong,Indonesia. These include the holotypes of the four new species, which were photographed before their preservation in 96% alcohol / 4% formalin solution.Sampling details, sex, and disk width (DW) were recorded (Borsa et al., 2016b). Tissue samples used for DNA extraction were deposited at LIPI-P2O in Ancol, Jakarta; a partial list of tissue samples is available from Arlyza et al. (2013a).
The phylogenetic tree of the blue-spotted maskray species complex (previouslyN.kuhlii), based on the concatenated partial nucleotide sequences of theCO1andcytochromebgenes showed nine main clades which characterizedN.australiae,N.caeruleopunctata,N.orientale,N.varidensand 5 other lineages that remained undescribed. The New Caledonian maskrayN.trigonoideswas considered as outgroup to the species complex (Borsa et al., 2013a). Voucher specimens deposited in museum collections are available for four of these lineages, i.e., cladesII,III,VIIandVIIIof Borsa et al. (2016b) (Fig.1b), but not yet for the fi fth (cladeI), herafter referred to as the Indian-Ocean maskray. Two additional lineages, each represented by a singleCO1haplotype (Supplementary Table S1) were not included in our phylogeny. In the following, they will be referred to as the Guadalcanal maskray and the Ryukyu maskray.
The partialCO1gene sequences here used to describe the holotypes of new species were 611 bp long. Variable nucleotide sites at theCO1locus were identified automatically using Mega6 (Tamura et al.,2013) on the matrix of 314 partialCO1gene sequences extracted from Borsa et al. (2016b). Diagnostic nucleotide sites were identified along a core 519-bp sub-fragment common to all 314 sequences by visual examination of the ExCel fi le (Microsoft Corporation,Redmond WA) with variable sites generated by Mega6 (Supplementary Table S1).Cytochromebgene sequences were not available for the type specimens of the new species except one: MZB-20864 from Ambon, to be chosen as paratype ofN.moluccensissp. nov. Variable nucleotide sites at thecytochromeblocus were similarly identified automatically using Mega6 (Tamura et al., 2013) on the matrix of 159 partialcytochromebgene sequences extracted from Borsa et al. (2016b). Diagnostic nucleotide sites were similarly identified along a core 1 127-bp sub-fragment sequenced in at least one individual of each species, by visual examination of the ExCel fi le with variable sites generated by Mega6(Supplementary Table S2).
The BOLD datasystems distinguishes clusters of sequences that qualify as operational taxonomic units(OTUs) i.e., putative species using the Refi ned Single Linkage (RESL) algorithm. The latter “clusters sequences with high similarity and connectivity and separates those with lower similarity and sparse connectivity” (Ratnasingham and Hebert, 2013).Each OTU thus identified is allocated a unique barcode index number (BIN) in BOLD. We established the homology of BIN numbers with the blue-spotted maskray lineages examined in the present study by visual inspection of the placement of the correspondingCO1gene sequences retrieved from BOLD in a maximum-likelihood tree of 314CO1gene sequences extracted from Borsa et al. (2016b), rooted byN.trigonoides. The tree was constructed with MEGA6(Tamura et al., 2013) using the Tamura 3-parameter model (Tamura, 1992) with gamma-distributed rate differences among sites, a choice based on the Bayesian information scores provided by MEGA6.
Notice: the present article in portable document(.pdf) format is a published work in the sense of the International Code of Zoological Nomenclature(International Commission on Zoological Nomenclature, 2012) or Code and hence the new names contained herein are effectively published under the Code. This published work and the nomenclatural acts it contains have been registered in ZooBank (http://zoobank.org/), the online registration system for the International Commission on Zoological Nomenclature. The ZooBank life science identifi er (LSID) for this publication is 97923E24-AD8A-4E72-978D-B691388AA503. The online version of this work is archived and available from theCJOLand haL-IRD repository (http://www.hal.ird.fr/) websites.
3 RESULT AND DISCUSSION
The way four species previously underN.kuhlii,namelyN.australiae,N.caeruleopunctata,N.orientale,N.varidenshave been diagnosed is not satisfactory (Table 1). Of all 19 characters used in the diagnoses of these four species, only one, the ratio of preoral length to mouth width, enabled the distinction ofN.kuhliifrom the other three species considered,but this was based on very low sample sizes (Table 1).Also, preoral length concerns a fl eshy part of the body whose shape is susceptible to be inf l uenced by environmental conditions during development and growth. Snout angle apparently enabled the distinction ofN.caeruleopunctataandN.orientalefromN.australiaeandN.kuhlii(Table 1), but the repeatability and precision of this measurement are likely to be poor, because of the subjectivity in defi ning linear sides to snout at its apex. The morphological key proposed by Last et al. (2016) was mostly based on the morphological traits of a few(1-10) individuals per species, with no attempt to assess presumed interSpecific differences against infra-Specific variation. The values of mostmorphological characters claimed to be diagnostic of a species overlapped across species (Table 1),suggesting that they are not suffcient to fully distinguish closely related, though genetically distinct species in the blue-spotted maskray species complex.Moreover, the apportion of environmental vs. genetic determination in the traits measured has not been evaluated. Therefore, the lack of diagnostic morphometric characters used for the “in-depth morphometric analyses” performed by Last et al.(2016) on the blue-spotted maskray species complex raises the question of their taxonomic utility. This question is a crucial one, not restricted to the bluespotted maskray species complex, because it has been claimed that these analyses follow “standard taxonomic practices used for the family group(stingrays)” (Last et al., 2016). We are here questioning the relevance of the morphometric diagnoses provided thus far in the blue-spotted maskray (Last et al.,2016), as in another species complexes in stingrays(Borsa, 2017). In contrast, we observe that mitochondrial-DNA sequences provide clear speciesSpecific clusters, offering much better diagnoses than so-called standard morphological measurements(Naylor et al., 2012; Puckridge et al., 2013; Borsa et al., 2016b; Last et al., 2016; Borsa, 2017). To eventually design a useful morphological key,multivariate analysis of morphometric characters should be undertaken posterior to molecular clustering, and the most discriminant of these characters, if any, should be selected. The discriminant characters can then be used for identifi cation purposes if no DNA barcoding is possible. This is beyond the scope of the present paper.
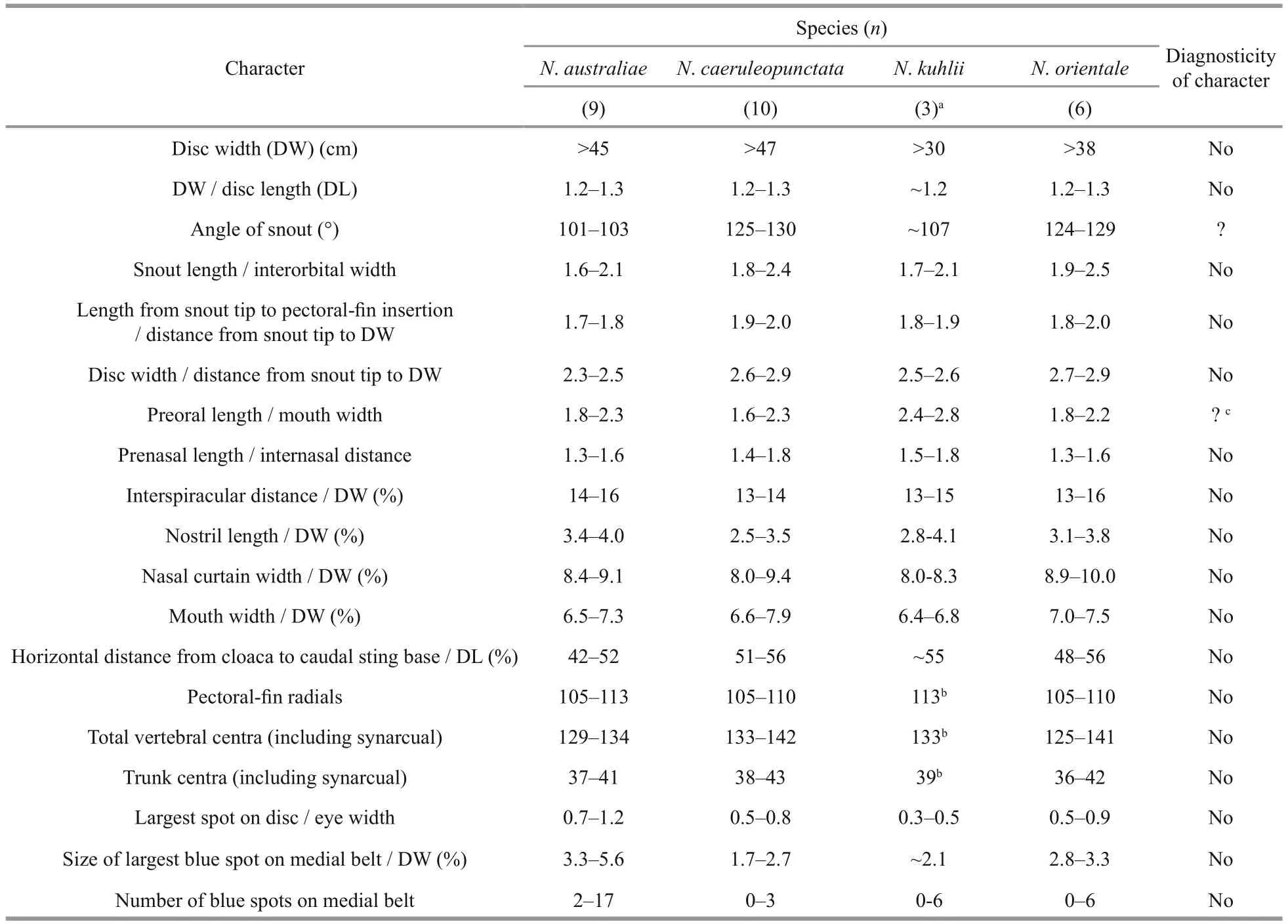
Table 1 Ranges of values for 19 morphological characters claimed to be diagnostic of four species in the blue-spotted maskray species complex (data compiled from Last et al., 2016)
Thus, the need arises for a molecular diagnosis of the species previously underN.kuhlii. Here, a 519-bp fragment of the nucleotide sequence of theCO1gene provided at least one, up to four diagnostic or quasidiagnostic nucleotides for eight (i.e.N.australiae,N.caeruleopunctata,N.varidens, cladesI,II,III,VII,VIII) out of the nine main lineages of blue-spotted maskray previously underN.kuhlii(Supplementary Table S1). Although no diagnostic nucleotide site was scored along this fragment for the remaining lineage(N.orientale), the latter was easily diagnosed by a combination of nucleotides at four nucleotide sites(see Taxonomy section). Two more lineages(Guadalcanal maskray, Ryukyu maskray), each represented by a single sequence were also taken into account for selecting diagnostic sites in the other lineages. Six of the nine blue-spotted maskray lineages treated in the present revision are currently represented in BOLD (see details in Taxonomy section). A distinct BIN number has been allocated to three of them (N.australiae,N.orientale,N.varidens)illustrating the taxonomic value of DNA barcodes in maskrays. However,N.caeruleopunctata, cladeIand cladeIIIwhich are reciprocally monophyletic (Borsa et al., 2016b) shared the same single BIN, exposing the limitations of the RESL analysis onCO1barcodes when addressing shallow divergences between clades,especially when sample sizes are low. It should be emphasized that BIN assignment on BOLD is a dynamic process, where individual BINs might split or merge each time new sequences are added(Ratnasingham and Hebert, 2013; Steinke et al.,2017). Similarly, a 1 127-bp fragment of the nucleotide sequence of thecytochromebgene provided at least one, up to nine diagnostic or quasidiagnostic nucleotides for eight (i.e.N.australiae,N.caeruleopunctata,N.varidens, cladesI,II,III,VII,VIII) out of the nine main lineages of blue-spotted maskray previously underN.kuhlii(Supplementary Table S2). Last, partial sequences of the nuclear recombination activating protein 1 (RAG-1) gene distinguish two main clades among species in the genusNeotrygon(Puckridge et al., 2013). One clade includes all the assayed sequences ofN.caeruleopunctata,N.trigonoides, cladeIand cladeIII; the other clade includes all the assayed sequences ofN.australiae,N.leylandi,N.picta,N.orientaleandN.varidens(see Fig.3B of Borsa et al., 2016b).
We recognize several cryptic species withinN.kuhliiunder its previous defi nition (Borsa et al.,2016b) and these species have parapatric distribution(Fig.1a). Given this, a specimen can also be identified to species from the locality where it has been collected,provided the locality of collection does not lie on the line of contact between geographically adjacent species. The samples examined by Borsa et al.(2016b) spanned the whole Indo-Malay-Papua archipelago, allowing us to map the geographic distribution of each of the eight lineages present in this region (Fig.1a) with higher accuracy than previous research (Arlyza et al., 2013a; Borsa et al.,2013a; Puckridge et al., 2013; Last et al., 2016).Geographic distributions are updated in the following Taxonomy section and previous speculative statements concerning the distribution ofN.australiae(Last et al., 2016) are also corrected.
The blue-spotted maskray is currently categorized as “data defi cient” by the International Union for the Conservation of Nature (Fahmi and Jacobsen, 2015).There are currently no species-Specific conservation measures in place for this species complex (Fahmi and Jacobsen, 2015). It is therefore urgent to have these species recognized and named, so appropriate species-Specific conservation measures can start to be implemented. The present descriptions are exclusively based on genetic diagnoses, unlike “standard”taxonomic descriptions in sharks and rays (White and Last, 2012; Last et al., 2016). The authors of recent“standard” descriptions in stingrays have claimed the new species were diagnosed based on morphology,but in some cases it can be inferred that the species were actually delineated from DNA sequences and not from morphology (see our present reassessment of Last et al., 2016; Table 1). The rationale for basing a new species description on genetic data has been repeatedly written in black ink (Tautz et al., 2003;Cook et al., 2010). In short, there is no objective reason to dismiss DNA-based descriptions when DNA sequences prove effective and suffcient as the only description of a species. New animal species have to be named and described according to the rules of zoological nomenclature (International Commission on Zoological Nomenclature, 1999),which at no moment specify which particular approaches, whether morphological, or genetical, or else, should be adopted or discarded to delineate and diagnose species. Indeed, the essence of the taxonomic description of a species is the science, and not the type of data on which it is based. Unless one arbitrarily imposed as the sole acceptable defi nition of a species the narrow morphological species concept,morphology thus should be considered as a mere possible source of taxonomic characters among many other potential sources (Cook et al., 2010). In invertebrates, species of the blue mussel have been re-described based on allozyme diagnoses (McDonald and Koehn, 1988), and the fi eld of entomology offers an emblematic example of DNA-only descriptions(Brower, 2010). In Actinopterygian fishes, new genera have been proposed based on DNA phylogenies exclusively (Craig and Hastings, 2007; Durand et al.,2012), and countless species have been described or resurrected based primarily on genetic markers (e.g.,Baldwin et al., 2011; Borsa et al., 2013c, 2014;Woodland and Anderson, 2014; Randall and Victor,2015; Shen et al., 2017). We believe that the present taxonomic revision will provide part of the information that is urgently needed for a sound management and conservation of cryptic species of the blue-spotted maskray in the Coral Triangle region.

Fig.1 Geographic structure of the blue-spotted maskray species complexa. geographic ranges of eight blue-spotted maskray species previously underNeotrygon kuhlii, including four new species [from the point map of Borsa et al.(2016b)] (dotted grey envelopes). Dotted purple ellipse:N.trigonoides. Circles indicate the sampling sites of three additional lineages [Guadalcanal maskray(G); Indian-Ocean maskray (I) and Ryukyu maskray (R)] and the type-locality ofN.kuhlii; b. simplified maximum-likelihood phylogenetic tree of bluespotted maskray species previously underN.kuhlii, including four new species, based on nucleotide sequences of the concatenated CO1+ cytochromebgene fragments (127 sequences, trimmed to a core length of 1 415 bp; Borsa et al., 2016b). New Caledonian maskrayN.trigonoideswas used as outgroup (Borsa et al., 2013a). Tree constructed under Mega6 (Tamura et al., 2013) (Tamura-Nei model with gamma-distributed rate differences among sites + invariant sites;partial deletion). Numbers at a node are bootstrap scores (from 600 bootstrap resampling runs). Roman numbers in brackets follow clade nomenclature of Arlyza et al. (2013a). No nucleotide sequence was available forN.kuhlii, which based on colour patterns and geographic proximity is likely a close relative ofN.trigonoidesif not synonymous with it (Borsa and Béarez, 2016).
4 TAXONOMY
Maskrays, genusNeotrygonCastelnau, 1873 belong to family Dasyatidae (Jordan, 1888). The type species of the genus isN.trigonoides(de Castelnau,1873), which has been recently resurrected from synonymy withN.kuhlii(Borsa et al., 2013a).
4.1 Description of four new maskray, Neotrygon spp. species previously under N. kuhlii
Neotrygon bobwardi sp. nov. http://zoobank.org/58F189B8-3504-46FD-B4DB-95CAE1712612.CladeII(Borsa et al., 2016b);NeotrygonkuhliicladeII(Arlyza et al., 2013a; Borsa et al., 2016a);Neotrygon kuhlii(Borsa et al., 2013a).
Typematerial: the holotype (Fig.2a) is a male specimen, 40 cm disk DW, collected by ISA on 28 April 2009 from local fi shermen at the Meulaboh,Aceh fi sh landing place (04°07′N 96°08′E). It is registered at MZB under No. MZB-20843; it is also registered at LIPI-P2O in Ancol, Jakarta under No.LIPI-4406. The specimen is preserved in 96% alcohol and 4% formalin mixture. A subsample of tissue,numbered NK-ME3 is currently preserved in alcohol at -20°C in the freezers of the genetics laboratory at LIPI-P2O. Paratypes are MZB-20842 (LIPI-4407),male, 32 cm DW from Pulau Breueh, Aceh, collected on 23 April 2009 by ISA; MZB-20844 (LIPI-4404),male, 20 cm DW from Sibolga, northwestern Sumatra,collected on 24 March 2009 by ISA; and MZB-20845(LIPI-4411) from Padang, West Sumatra, collected in August 2009 by ISA.

Fig.2 Holotypes of four new maskray species previously under Neotrygon kuhlii (Müller and Henle, 1841)a.Neotrygon bobwardisp. nov., No. MZB-20843 from Meulaboh, Aceh; male, 40 cm DW (photograph by ISA); b.Neotrygon malaccensissp. nov., No.MZB-20847 from Kuala Lama, Malacca Strait; female, 40 cm DW (photograph by ISA); c.Neotrygon moluccensissp. nov., No. MZB-20866 from Tual, Kei Islands; female, 30.5 cm DW; d.Neotrygon westpapuensissp. nov., No. MZB-20867 from Biak, West Papua; female, 36 cm DW.
Description: the partialCO1gene sequence of the holotype, which has GenBank accession No.JX304805, is 5′- C T G G C C T C A G T T T A C T T A T C C G A A C A G A A C T A A G C C A A C C A G G C G C T T T A C T G G G T G A T G A T C A A A T T T A T A A T G T A A T C G T C A C T G C C C A C G C C T T C G T A A T A A T C T T C T T T A T G G T A A T G C C A A T T A T A A T T G G T G G G T T T G G C A A C T G A C T G G T G C C C C T G A T A A T T G G G G C T C C G G A C A T A G C C T T T C C A C G A A T A A A C A A C A T A A G T T T T T G A C T T C T A C C T C C C T C A T T C C T A T T A C T G C T G G C C T C A G C A G G A G T A G A A G C C G G A G C C G G A A C A G G T T G A A C A G T T T A T C C C C C A T T A G C T G G T A A T C T A G C A C A T G C C G G A G C T T C T G T A G A C C T T A C A A T C T T C T C T C T T C A C C T A G C A G G T G T T T C C T C T A T T C T G G C A T C C A T C A A C T T T A T C A C A A C A A T T A T T A A T A T A A A A C C A C C T G C A A T C T C C C A G T A T C A A A C C C C A T T A T T C G T C T G A T C T A T T C T T G T T A C A A C T G T G C T T C T C C T G C T A T C T C T A C C A G T C C T A G C A G C T G G C A T T A C C A T A C T C C T C A C A G A C C G A A A T C T T A A T A C A A C T T T C T T T G A C C C A G C T G G G G G A G G A G A T C C C A T T C T T T A C C A A C A C C T C -3′.
Diagnosis: based on Supplementary Table S1,N.bobwardisp. nov. is distinguished from all other species currently underN.kuhliiby nucleotide G at nucleotide site 210 of theCO1gene; it also has C at nucleotide site 240, a character that it shares with no species other thanN.malaccensissp. nov. Based on Supplementary Table S2,N.bobwardisp. nov. is distinguished from other species previously underN.kuhliiby C at nucleotide site 546 of thecytochrome bgene; it also has C at nucleotide site 399, a character that it shares with no species other thanN.malaccensissp. nov., G at nucleotide 57 and T at nucleotide site 111, characters that it shares with no species other thanN.caeruleopunctatasp. nov., and T at nucleotide site 327, a character that it shares with no species other thanN.westpapuensissp. nov.
Distribution: the type locality ofN.bobwardisp.nov. is Meulaboh, northwestern coast of Sumatra Island. Based on present study, its distribution includes the northern tip of the Aceh region and all the western coast of Sumatra south to Padang.
Etymology: the species is named after Robert D.(Bob) Ward, one of the leaders of the fi sh barcoding initiative (Ward et al., 2009). One of his noted contributions in this fi eld was the DNA-barcoding survey of Australian chondrichthyans, which included bluespotted maskray samples from the Coral Triangle region (Ward et al., 2008). R. D. Ward and co-authors suspected the occurrence of cryptic species within the nominal speciesN.kuhlii, based on an unusually high level of genetic divergence among geographic populations, at theCO1locus. We chose to name after him the bluespotted maskray species that occurs on the Indian-Ocean coast of Sumatra, as an acknowledgement of his contribution to the systematics of chondrichthyans.
Proposedvernacularnames: Bob Ward’s bluespotted maskray (English); pari masker totol biru Pak Bob (Indonesian); raie pastenague à points bleus de Bob Ward (French).
Neotrygon malaccensis sp. nov. http://zoobank.org/69B1F017-99AD-4E5E-81BE-2684E2192D1A.CladeIII(Borsa et al., 2016b);NeotrygonkuhliicladeIII(Arlyza et al., 2013a; Borsa et al., 2016a);Neotrygonkuhliiclade7(Puckridge et al., 2013);Neotrygonkuhlii(Borsa et al., 2013a). Also BIN number BOLD:AAA5611 in BOLD.
Typematerial: the holotype (Fig.2b) is a female specimen, 40 cm DW, collected by ISA on 06 December 2008 at the fi sh landing site of Kuala Lama,northeastern Sumatra Island (03°26′N, 99°16′E). It is registered at the Museum Zoologicum Bogoriense under no. MZB-20847 (one of two specimens); it is also registered at LIPI-P2O under No. LIPI-4401 (one of two specimens). Whole specimen preserved in 96% alcohol and 4% formalin. A subsample of tissue,designated NK-MSKL3 is currently preserved in alcohol at -20°C in the freezers of the genetics laboratory at LIPI-P2O. The single paratype is the larger specimen under registration No. MZB-20847(LIPI-4401; tissue sample no. NK-MSKL4); this is a female, 42 cm DW, from Kuala Lama, collected in March 2009 by ISA.
Description: the partialCO1gene sequence of the holotype, which has GenBank accession No.JX304818, is 5′- C T G G C C T C A G T T T A C T T A T C C G A A C A G A A C T G A G C C A A C C A G G C G C T T T A C T G G G T G A T G A T C A A A T T T A T A A T G T A A T C G T C A C T G C C C A C G C C T T C G T A A T A A T C T T C T T T A T A G T A A T G C C A A T T A T A A T T G G T G G G T T T G G C A A C T G A C T A G T G C C C C T G A T A A T T G G G G C T C C G G A C A T A G C C T T T C C A C G A A T A A A C A A C A T A A G T T T T T G A C T T C T A C C T C C C T C A T T C C T A T T A C T G C T A G C C T C A G C A G G A G T A G A A G C C G G A G C C G G A A C A G G T T G A A C A G T T T A T C C T C C A T T A G C T G G T A A T C T A G C A C A T G C C G G A G C T T C T G T A G A C C T T A C A A T C T T C T C T C T T C A C C T A G C A G G T G T T T C C T C T A T T T T G G C A T C C A T C A A C T T T A T C A C A A C A A T T A T T A A T A T A A A A C C A C C T G C A A T C T C C C A A T A T C A A A C C C C A T T A T T C G T C T G A T C T A T T C T T G T T A C A A C T G T A C T T C T C C T G C T A T C C C T A C C A G T C C T A G C A G C T G G C A T T A C T A T A C T C C T C A C A G A C C G A A A T C T T A A T A C A A C T T T C T T T G A C C C A G C T G G G G G A G G A G A T C C C A T T C T T T A C C A A C A C C T C -3′.
Diagnosis: based on Supplementary Table S1,N.malaccensissp. nov. is distinguished from other species previously underN.kuhliiby G at nucleotide site 126 of theCO1gene. MostN.malaccensissp.nov. individuals have T at nucleotide site 478, a character that was otherwise present in a singleN.orientaleindividual;N.malaccensissp. nov. also has C at nucleotide site 240, a character that it shares exclusively withN.bobwardisp. nov. and T at nucleotide site 393, a character that it shares exclusively with the Guadalcanal maskray. Based on Supplementary Table S2,N.malaccensissp. nov. is distinguished from other species previously underN.kuhliiby C at nucleotide sites 129 and 441, G at nucleotide site 405, and T at nucleotide site 264 of thecytochromebgene; T at nucleotide site 225 is quasidiagnostic; it also has C at nucleotide 399, a character that it shares with no species other thanN.bobwardisp. nov. Based on Puckridge et al. (2013), partial haplotypes at theRAG-1locus enable the distinction ofN.malaccensissp. nov. fromN.australiae,N.orientaleandN.varidens, but not from eitherN.caeruleopunctata,N.trigonoidesor the Indian-Ocean maskray.
Distribution: the type locality ofN.malaccensissp. nov. is Kuala Lama, Malacca Strait (03°39′N,98°59′E). Specimens of the same species were sampled from Perbaungan, a neighbouring locality in the Malacca Strait and from the western coast of the Kra isthmus, Thailand. Known distribution, based on present study: northern part of Malacca Strait and eastern Andaman Sea.
Etymology: epithetmalaccensisrefers to the geographic origin of the type material, the Malacca Strait. It is the latinized geographical adjectival form of noun “Malacca”.
Proposedvernacularnames: the Malacca Strait bluespotted maskray (English); pari masker totol biru Selat Melaka (Indonesian); raie pastenague masquée à points bleus du détroit de Malacca (French).
Neotrygon moluccensis sp. nov. http://zoobank.org/626567B0-3F01-4233-B7CA-2F617DE9186B.CladeVII(Borsa et al., 2016b);NeotrygonkuhliiCladeVII(Arlyza et al., 2013a; Borsa et al., 2016a);Neotrygonkuhlii(Borsa et al., 2013a).
Typematerial: the holotype (Fig.2c) is a female specimen, 30.5 cm DW, collected by A. Kusnadi on 08 April 2009 in front of the LIPI laboratories in Tual,Kei Islands (05°38′S, 132°44′E). It is registered at the Museum Zoologicum Bogoriense under No. MZB-20866; also registered at LIPI-P2O under No. LIPI-4405. Preserved in 96% alcohol and 4% formalin. A subsample of tissue, numbered ARA1 is currently preserved in alcohol at -20°C in the freezers of the genetics laboratory at LIPI-P2O. The single paratype is MZB-20864 (LIPI-4400), female, 36 cm DW from Ambon Bay, collected on 23 October 2008 by ISA.
Description: the partialCO1gene sequence of the holotype, which has GenBank accession no.JX304898, is 5′- C T G T T C T T A G T T T A C T T A T C C G A A C A G A A C T A A C C C C A C C A G G C G C T T T A C T G G G T G A T G A T C A A A T T T A T A A T G T A A T C G T C A C T G C C C A C G C C T T C G T A A T A A T C T T C T T T A T A G T A A T G C C A A T T A T A A T T G G T G G G T T T G G T A A T T G A C T A G T G C C C C T G A T G A T T G G G G C T C C G G A C A T A G C C T T T C C A C G A A T A A A C A A C A T A A G T T T C T G A C T T C T A C C T C C C T C A T T C C T A T T A C T G C T A G C C T C A G C A G G A G T A G A A G C C G G A C C T G G G A C A G G T T G A A C A G T T T A T C C C C C A T T A G C T G G T A A T C T A C C A C A T G C C G G A G C T T C T G T A G A C C T T A C A A T C T T T T C T C T T C A C C T A G C A G G T G T T T C C T C T A T T C T A G C A T C C A T C A A C T T T A T C A C A A C A A T T A T T A A T A T A A A A C C A C C T G C A A T C T C C C A G T A T C A A A C C C C A T T A T T C G T C T G A T C T A T T C T T G T T A C A A C T G T A C T T C T C C T G C T A T C C C T A C C A G T C C T A G C A G C T G G C A T T A C T A T A C T C C T C A C A G A C C G A A A T C T T A A T A C A A C T T T C T T T G A C C C A G C T G G A G G A G G A G A T C C C A T T C T T T A C C A A C A C C T C -3′.
Diagnosis: based on Supplementary Table S1,N.moluccensissp. nov. is distinguished from other species previously underN.kuhliiby T at nucleotide site 447 of theCO1gene;N.moluccensissp. nov.also has G at nucleotide site 261, a character that it shares with the Guadalcanal maskray and with a singleN.australiaeindividual, and C at nucleotide site 309, a character that it shares with the Guadalcanal maskray and with a few individuals from the Indian Ocean. Based on Supplementary Table S2,N.moluccensissp. nov. is distinguished from other species previously underN.kuhliiby C at nucleotide site 519, G at nucleotide site 453, and T at nucleotide sites 291, 906 and 1125 of thecytochromebgene; it also has C at nucleotide site 246, a character that it shares with no species other thanN.caeruleopunctata,and G at nucleotide site 1116, a character that it shares with no lineage other than the Indian-Ocean maskray.
Distribution: the type locality ofN.moluccensissp. nov. is Tual, Kei Islands, Moluccas, Indonesia.Specimens of the same species were sampled from Ambon, Moluccas. Known distribution, based on present study: eastern half of Banda Sea.
Etymology: epithetmoluccensisrefers to the geographic origin of the type material, the Molucca islands. It is the latinized geographical adjectival form of noun “Molucca”.
Proposedvernacularnames: Moluccan bluespotted maskray (English); pari masker totol biru Maluku(Indonesian); raie pastenague masquée à points bleus des Moluques (French).
Neotrygon westpapuensis sp. nov. http://zoobank.org/565C798C-249E-4BA9-9B02-ED2189C33FA2.CladeVIII(Borsa et al., 2016b);NeotrygonkuhliicladeVIII(Arlyza et al., 2013a; Borsa et al., 2016a);Neotrygonkuhlii(Borsa et al., 2013a).
Typematerial: the holotype (Fig.2d) is a female specimen, 36 cm DW, collected by Alvi Sitepu on 06 June 2009 in Biak, West Papua. It is registered at the Museum Zoologicum Bogoriense under No. MZB-20867; it is also registered at LIPI-P2O under no.LIPI 4408. The specimen is preserved in 96% alcohol and 4% formalin mixture. A subsample of tissue,numbered NK-BK5 is currently preserved in alcohol at -20°C in the freezers of the genetics laboratory at LIPI-P2O.
Description: the partialCO1gene sequence of the holotype, which has GenBank accession No.JX304909 is 5′- C T G G C C T C A G T T T A C T T A T C C G A A C A G A A C T A A G C C A A C C A G G C G C T T T A C T G G G T G A T G A T C A A A T T T A T A A T G T A A T C G T C A C T G C C C A C G C C T T C G T A A T A A T C T T C T T T A T A G T A A T G C C A A T T A T A A T T G G T G G G T T T G G T A A T T G A C T A G T A C C C C T A A T A A T T G G G G C T C C G G A C A T A G C C T T T C C A C G A A T G A A C A A C A T A A G T T T T T G A C T T C T G C C T C C C T C A T T C C T A C T A C T G C T A G C C T C A G C A G G G G T A G A A G C C G G A G C C G G A A C A G G T T G A A C A G T T T A T C C C C C A T T A G C T G G T A A T C T A G C A C A T G C C G G A G C T T C T G T A G A C C T T A C A A T C T T C T C T C T T C A C C T A G C A G G T G T T T C C T C T A T T C T G G C A T C C A T C A A C T T T A T C A C A A C A A T T A T T A A T A T A A A A C C A C C T G C A A T C T C C C A G T A T C A A A C C C C A T T A T T C G T C T G A T C T A T T C T T G T T A C A A C T G T A C T T C T C C T G C T A T C C C T A C C A G T C C T A G C A G C T G G C A T T A C T A T A C T C C T C A C A G A C C G A A A T C T T A A T A C A A C T T T C T T T G A C C C A G C T G G A G G G G G A G A T C C C A T T C T T T A C C A A C A C C T C -3′.
Diagnosis: based on Supplementary Table S1,N.westpapuensissp. nov. is distinguished from all other species previously underN.kuhliiby G at nucleotide 354 of theCO1gene; it also possesses G at nucleotide site 294, a character that it shares with no other species thanN.caeruleopunctataand C at nucleotide site 334, a character that is otherwise present in the Ryukyu maskray only. Based on Supplementary Table S2,N.westpapuensissp. nov. is distinguished from other species previously underN.kuhliiby C at nucleotide site 733, G at nucleotide sites 76, 372, 528 and 753, and T at nucleotide sites 48, 57, 228 and 792 of thecytochromebgene; it also has T at nucleotide site 327, a character that it shares with no species other thanN.bobwardisp. nov.
Distribution: the type locality ofN.westpapuensissp. nov. is Biak Island north of Cenderawasih Bay,West Papua. As all samples we have of this species originate from Biak and from nearby Numfor Island only, amid a wide unsampled region (Fig.1a), no distribution map can be inferred at present. The geological features of the northern coastline of the Vogelkop peninsula, where the shallow-water habitat favourabe to blue-spotted maskrays is absent, isolate the reef fauna of Cenderawasih Bay from that of western West Papua further west. Further research is needed to delineate the precise geographic distribution ofN.westpapuensissp. nov.
Etymology: Named for the country of type locality,West Papua as it is spelled since 1961 (Saltford,2003). Epithetwestpapuensisis the latinized geographical adjectival form of noun “West Papua”,with no emendation.
Proposedvernacularnames: West Papuan bluespotted maskray (English); pari masker totol biru West Papua (Indonesian); raie pastenague masquée à points bleus de Papouasie Occidentale (French).
4.2 Molecular diagnoses and updated distributions of N. australiae, N. caeruleopunctata, N. orientale and N. varidens
Neotrygon australiae Last et al., 2016.Neotrygon kuhliicladeV(Arlyza et al., 2013a; Borsa et al.,2016a);Neotrygonkuhliiclade5(Puckridge et al.,2013); cladeNeotrygonkuhlii4of Naylor et al.(2012);Neotrygonkuhlii(Cerutti-Pereyra et al., 2012;Borsa et al., 2013a). Also BIN number BOLD:AAA5609 in BOLD.
Diagnosis: based on Supplementary Table S1,N.australiaeis distinguished from other species previously underN.kuhliiby T at nucleotide sites 124, 171 and 435 of theCO1gene; C at nucleotide site 321 is quasi-diagnostic. Based on Supplementary Table S2,N.australiaeis distinguished from other species previously underN.kuhliiby A at nucleotide site 750, C at nucleotide sites 556 and 831, and T at nucleotide site 705 of thecytochromebgene; T at nucleotide site 642 is partly diagnostic. Based on Puckridge et al. (2013), partial haplotypes at theRAG-1locus enable the distinction ofN.australiaefromN.caeruleopunctata,N.malaccensissp. nov.,N.trigonoidesand the Indian-Ocean maskray, but not from eitherN.orientaleorN.varidens.
Distribution: the type locality ofN.australiaeis southwest of Weipa, Gulf of Carpentaria (Last et al.,2016). Known distribution, based on Borsa et al.(2016b) (Fig.1a): Timor Island, Rote Island, Lesser Sunda islands including Lombok and Flores, Sahul shelf, from Ningaloo Reef to Torres Strait and the northern extremity of the Great Barrier Reef. Last and co-authors wrote that the distribution ofN.australiaealso encompassed eastern Indonesia, West Papua and New Guinea, but this is totally unwarranted. Not only did these authors examine no blue-spotted maskray sample from these regions, but previous research had shown that the blue-spotted maskray samples from Ambon and Kei Islands in the Moluccas and from Biak Island in West Papua corresponded to distinct mitochondrial clades, themselves distinct from the clade that characterizesN.australiae(Arlyza et al.,2013a; Borsa et al., 2016a).
Remarks: Last et al. (2016) assigned Müller and Henle’s syntype from New Guinea (MNHNIC-A-7931; collected by J. R. C. Quoy and J. P.Gaimard during the hydrographic expedition of theAstrolabe, 1826-1829) toN.australiae, without justifi cation. They placed the sampling locality of that specimen in “West Papua, New Guinea, 7°30′S,132°30′E”. These coordinates actually designate a location in the deep ocean 64 km east-southeast of the eastern tip of Larat Island in the Tanimbar archipelago,Moluccas. Not only the habitat at this location is unsuitable toN.kuhlii, which is a demersal, shallowwater chondrichthyan, but there is no indication from the collectors’ notes (Quoy and Gaimard, 1835;Bauchot, 1994) or from the expedition commander’s logbook (Dumont d’Urville, 1833) that the Tanimbar archipelago was ever visited by theAstrolabe. It has been instead determined that the New Guinea syntype originates from Manokwari in northern West Papua(00°52′S, 134°04′E) (Borsa and Béarez, 2016). Based on sampling location, specimen MNHN-IC-A-7931 more likely belongs toN.westpapuensissp. nov. thanN.australiae. Sequencing fresh specimens from Manokwari would have helped verify this but so far we only accessed specimens from Pulau Numfoor(Borsa et al., 2016b), ca. 50 km east of Manokwari Bay.
Neotrygon caeruleopunctata Last et al., 2016.NeotrygonkuhliicladeVI(Arlyza et al., 2013a; Borsa et al., 2016a);Neotrygonkuhliiclade6(Puckridge et al., 2013);Neotrygonkuhlii(Ward et al., 2008; Borsa et al., 2013a). Also BIN number BOLD:AAA5611 in BOLD.
Diagnosis: based on Supplementary Table S1,N.caeruleopunctatais distinguished from other species currently underN.kuhliiby the simultaneous possession of T at nucleotide site 457 of theCO1gene;N.caeruleopunctataalso has G at nucleotide site 294, a character that it shares exclusively withN.westpapuensissp. nov., C at nucleotide site 270, a character that it shares exclusively with a proportion ofN.orientaleindividuals, and C at nucleotide site 567, a character that it shares exclusively with the Guadalcanal maskray. Based on Supplementary Table S2,N.caeruleopunctatais distinguished from other species previously underN.kuhliiby C at nucleotide sites 513 of thecytochromebgene; the quartet (G, T,C, C) at nucleotide sites (57, 111, 327, 399) is also diagnostic. Based on Puckridge et al. (2013), partial haplotypes at theRAG-1locus enable the distinction ofN.caeruleopunctatafromN.australiae,N.orientaleandN.varidens, but not from eitherN.malaccensissp. nov.,N.trigonoidesor the Indian-Ocean maskray.
Distribution: the type locality ofN.caeruleopunctatais Kedonganan Bay, Bali, Indonesia.AllN.caeruleopunctataspecimens sampled thus far originated from the Indian-Ocean coasts of Java and Bali (Fig.1a). FourN.caeruleopunctataspecimens included in Puckridge et al. (2013), whose partialCO1gene sequence was identical to the holotype of the species were reportedly sampled in Sadang, an inland locality of Central Java. However, fi gure 3 of Puckridge et al. (2013) suggests that the sample originated from the southern coast of Java Island.
Neotrygon orientale Last et al., 2016.Neotrygon kuhliicladeIV(Arlyza et al., 2013a; Borsa et al.,2016a);Neotrygonkuhliiclades2and3(Puckridge et al., 2013); cladeNeotrygonkuhlii1of Naylor et al.(2012);Neotrygonkuhlii(Ward et al., 2008; Last et al., 2010; Aschliman et al., 2012; Borsa et al., 2013a;Shen et al., 2016). Also BIN number BOLD:ABZ6131 in BOLD.
Diagnosis: based on Supplementary Table S1,N.orientaleis distinguished from other species previously underN.kuhliiby having a combination of C at nucleotide site 228 of theCO1gene that it shares exclusively withN.varidensand the Guadalcanal maskray, T at nucleotide site 405, which excludesN.varidensand the pair (C, A) at nucleotide sites (420,522), which excludes the Guadalcanal maskray.Based on Supplementary Table S2,N.orientalehas C at nucleotide site 255 of thecytochromebgene, a character that it shares with no species other thanN.varidens. Based on Puckridge et al. (2013), partial haplotypes at theRAG-1locus enable the distinction ofN.orientalefromN.caeruleopunctata,N.malaccensissp. nov.,N.trigonoidesand the Indian-Ocean maskray, but not from eitherN.australiaeorN.varidens.
Distribution: the type locality ofN.orientaleis the eastern Java Sea off the southeastern tip of Borneo(Last et al., 2016). Based on the present study, its wide distribution includes the South China Sea, the Java Sea, the Sulu Sea, the Sulawesi Sea, the Philippines and Sulawesi Island (Fig.1a). Also sampled from Tanjung Luar, but exact geographic origin unknown (see Borsa et al., 2013b: 237).
Neotrygon varidens (Garman, 1885).Neotrygon varidens(Last et al., 2016);NeotrygonkuhliicladeIV(Arlyza et al., 2013a; Borsa et al., 2016a);Neotrygon kuhliiclade1(Puckridge et al., 2013); cladeNeotrygon kuhlii2of Naylor et al. (2012);Neotrygonkuhlii(Ward et al., 2008; Cerutti-Pereyra et al., 2012; Wang et al., 2012; Arlyza et al., 2013b, Borsa et al., 2013a;Chen et al., 2014);Dasybatusvaridens, original combination (Garman, 1885). Also BIN number BOLD:ABZ6130 in BOLD.
Diagnosis: based on Supplementary Table S1,N.varidensis diagnosed relative to other species previously underN.kuhliiby (C, T) at nucleotide sites (405, 456) of theCO1gene;N.varidensalso has C at nucleotide site 507, which it shares exclusively with a proportion ofN.bobwardisp. nov. individuals,and C at nucleotide site 228, which it shares exclusively withN.orientaleand the Guadalcanal maskray. Based on Supplementary Table S2,N.varidensis unique by having A at nucleotide site 480, C at nucleotide site 120, and T at nucleotide site 402 of thecytochromebgene; it also has C at nucleotide site 255, a character that it shares with no species other thanN.orientale, and T at nucleotide site 973, a character that it shares with no species other thanN.bobwardisp. nov. Based on Puckridge et al. (2013), partial haplotypes at the nuclear recombination activating protein 1 (RAG-1) locus enable the distinction ofN.varidensfromN.caeruleopunctata,N.malaccensissp. nov.,N.trigonoidesand the Indian-Ocean maskray, but not from eitherN.australiaeorN.orientale.
Distribution: the type locality ofN.varidensis Hong Kong (Garman, 1885). Its known distribution,based on Borsa et al. (2016b) is the South China Sea.
4.3 Other Neotrygon spp. lineages requiring further attention
Guadalcanal maskray.Guadalcanal maskray(Borsa et al., 2016b);Neotrygonkuhlii(Last et al.,2016). The single vouchered specimen available is CSIRO H7723-01 from Honiara, Guadalcanal,Solomon Islands. Its partialCO1gene sequence,which is unique among blue-spotted maskays has been published (Last et al., 2016). The limited morphological data available show that the Guadalcanal maskray is a species different fromN.kuhlii(Borsa and Béarez, 2016).
Indian-Ocean maskray.CladeI(Borsa et al.,2016b);NeotrygonkuhliihaplogroupI(Arlyza et al.,2013a; Borsa et al., 2016a);Neotrygonkuhliiclade8(Puckridge et al., 2013); presumably cladeNeotrygon kuhlii3of Naylor et al. (2012);Neotrygonkuhlii(Borsa et al., 2013a). Also BIN number BOLD:AAA5611 in BOLD. Voucher material for this clade includes a specimen collected by A. Pavan Kumar and colleagues on 15 August 2011 in Visakhapatnam, Andhra Pradesh state ofindia, Bay of Bengal. The specimen has been discarded (A.Pavan Kumar, pers. comm.) but a sub-sample of tissue has been registered at the Central Institute of Fisheries Education of Visakhapatnam under no.VIZNK-01. Its partialCO1gene sequence has GenBank accession no. JX978329. A photograph of this individual is available from BOLD under accession no. BOLD:ACB9305. Most (12/14) Indian Ocean maskray individuals in our dataset possessed T at nucleotide site 607 of theCO1gene, a character that was otherwise present in only 2/130N.orientaleindividuals (Supplementary Table S1). From the material genetically examined thus far, the Indian bluespotted maskray’s distribution includes the Indian coast of the Bay of Bengal (Visakhapatnam, Chennai),the Indian coast of the Laccadives Sea (Kerala), and eastern Africa (Borsa et al., 2016b).
Ryukyu maskray.Ryukyu maskray (Borsa et al.,2016b);NeotrygonkuhliicladeIV(Arlyza et al.,2013a; Borsa et al., 2016a);Neotrygonkuhliiclade4(Puckridge et al., 2013);Neotrygonkuhlii(Borsa et al., 2013a; Yagishita et al., 2009). Also BIN number BOLD:ACH3785 in BOLD. If more material confi rms that the Ryukyu maskray is genetically distinct from other blue-spotted maskrays, then this should be erected as a new species. The Ryukyu maskray has been reported from Ishigaki-shima; it was represented by a single nucleotide sequence (Yagishita et al.,2009) in our dataset.
5 DATA AVAILABILITY
All nucleotide sequences referred to in the present work have been deposited in GenBank (https://www.nlm.ncbi.nih.gov/). New species names have been registered in ZooBank (http://zoobank.org/). The holotypes of the four new species described in this paper have been deposited at MZB, Cibinong,Indonesia.
6 ACKNOWLEDGMENT
This paper is a contribution of a collaborative project on the population genetics of stingrays in the Indonesian archipelago (PARI), run jointly by IRD UMR 250 and LIPI-P2O. We thank R. K. Hadiaty(LIPI, Cibinong) for allocating MZB collection numbers to the holotypes and paratypes of the four new species described in the present paper. We had valuable discussions with P. Béarez (MNHN, Paris),P. Berrebi (CNRS, Montpellier), W.-J. Chen (NTU,Taipei), J.-D. Durand (IRD, Montpellier), N. Hubert(IRD, Cibinong) and R. D. Ward (CSIRO, Hobart).We are also grateful to two anonymous reviewers for insightful comments. We took criticism from P. Last(CSIRO, Hobart), B. Séret (MNHN, Paris), S.
Weigmann (Elasmo-Lab, Lüneburg) and W. T. White(CSIRO, Hobart) as encouragement. Our background map of the Indo-West Pacific was edited from images downloaded from Digital Vector Maps, San Diego(http://digital-vector-maps.com/). Several books including J. Dumont d’Urville’sVoyagedel’Astrolabeand J. Müller and F. G. J. Henle’sSystematischeBeschreibungderPlagiostomenwere consulted from the Biodiversity Heritage Library website (http://www.biodiversitylibrary.org). Designed the study:PB. Contributed materials or data or analysis tools:ISA, PB, TBH, KNS. Wrote the paper: PB. The authors declare no fi nancial conf l ict ofinterest.
杂志排行
Journal of Oceanology and Limnology的其它文章
- Response of the North Pacific Oscillation to global warming in the models of the Intergovernmental Panel on Climate Change Fourth Assessment Report*
- Effect of mesoscale wind stress-SST coupling on the Kuroshio extension jet*
- Surface diurnal warming in the East China Sea derived from satellite remote sensing*
- Cross-shelf transport induced by coastal trapped waves along the coast of East China Sea*
- Observations of near-inertial waves induced by parametric subharmonic instability*
- Seasonal variation and modal content ofinternal tides in the northern South China Sea*
