Alternate phenotype–genotype selection for developing superior high-yielding irrigated rice lines
2018-04-12YonnelleDeMoukoumbiRftElNmkyKoffiDjmnDoudMbodjBboucrrMnneh
Yonnelle De Moukoumbi*,Rft El-Nmky,Koffi Djmn,Doud Mbodj,Bboucrr Mnneh
a Irrigated Rice Breeding Unit,Africa Rice Center(AfricaRice),Sahel Regional Station,BP 96 Saint Louis,Senegal
b National Institute of Agricultural Research,Gros bouquet,PMB 16169,Libreville,Gabon
c Rice Research&Training Center(RRTC),33717 Sakha Kafr Sheikh,Egypt
1.Introduction
Rice is the second most important cereal crop in the world after maize in terms of cultivated area,with 158.8 Mha under production in 2016[1].Global paddy rice production was 2.9 Mt.to a record of 749.7 Mt.(497.9 Mt.on a milled basis).In Africa,the expected 2016 production was 29.7 Mt.(19.4 Mt.,milled basis),implying a 4%year-on-year expansion and a new record[2].At the Yield Potential International Workshop held by the Global Rice Science Partnership(GRiSP)in 2011,it was asserted that worldwide demand for rice is expected to rise by>25%by 2035[3].Since the 1960s,many high-yielding rice varieties and breeding lines have been developed by the International Rice Research Institute,including Oryza sativa L.IR8,IR36,IR64,and IR72[4].During the 1990s,Africa Rice Center(AfricaRice)scientists developed high-yielding upland and lowland New Rice for Africa(NERICA)and irrigated Sahel varieties[5,6]which have been distributed to farmers and breeders worldwide.
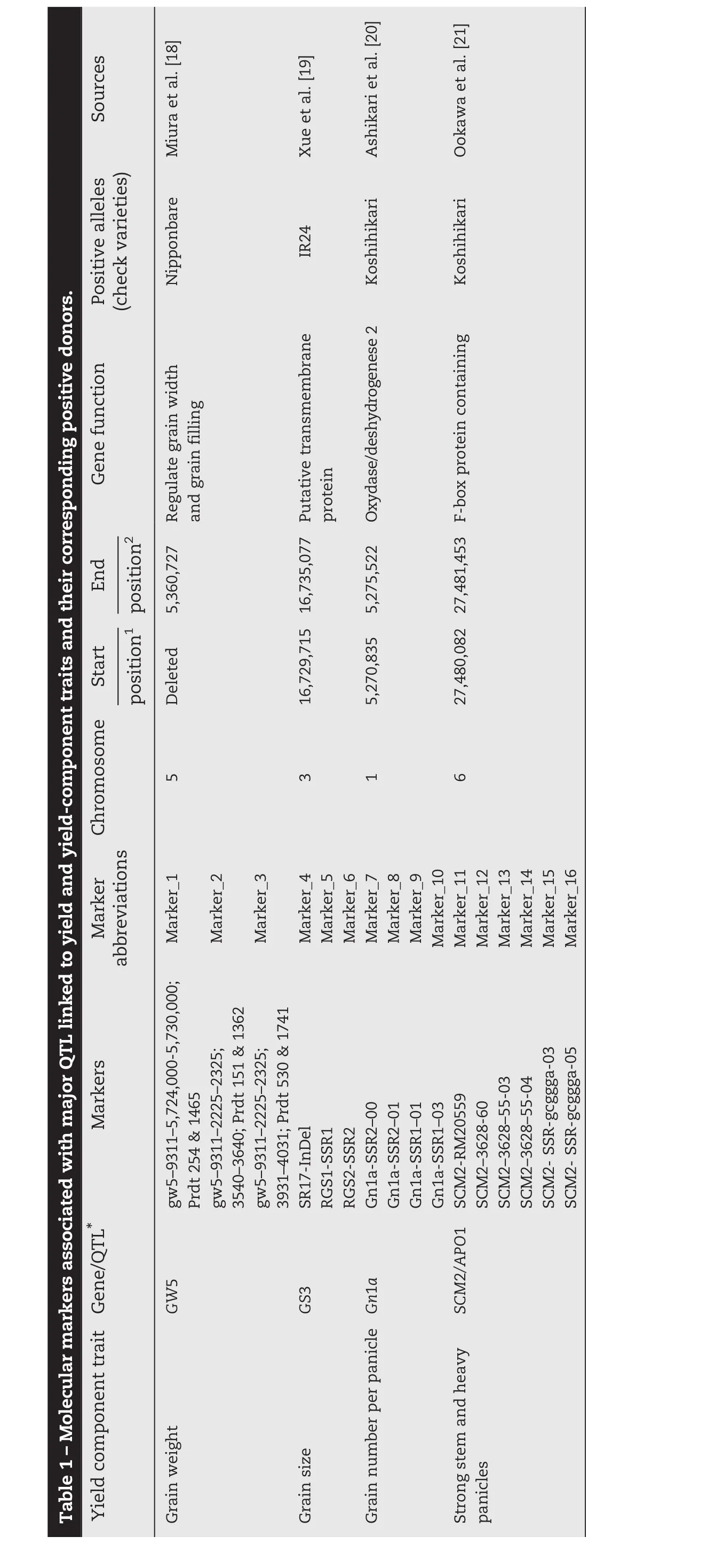
Yield potential is defined as the maximum achievable yield in the absence of biophysical,physiological,or economic constraints on production[7].Increasing rice yield potential is one of the most important contributions for any rice breeding program aimed at developing high-yielding varieties.High-yielding technologies that have been developed include “new plant type”, “hybrid rice”,and “super hybrid rice”adapted to specific cropping conditions[8–10].Rice research in Egypt during the past 15 years has increased the national average yield by>66%,from 5.71 to 9.84 t ha−1[11].This increase was achieved by growing modern inbred varieties,which cover almost 100%of the total rice area in the country.In West Africa,the current average yield potential of irrigated rice varieties such as the widely grown Sahel varieties developed by pedigree selection ranges from 10 to 12 t ha−1[12].Increasing yield potential requires continuous phenotypic selection of desirable lines from a large number of segregating populations until fixation of the desired trait[13,14].The numbers of plants to select at each generation may be modified according to the species,the breeding objective,and the genetics of the traits of interest.This method is labor-intensive and time-consuming and requires a large nursery or field space for screening.In the last decade,different approaches including the use of wide crosses and gene pyramiding through molecular approaches[15]have been used to improve rice yield potential.Physiological approaches using simulation models predicted that an increase in rice yield potential of 25%is possible by changing the traits of the current plant type[16].Molecular techniques are continuously being used to increase the number of genes discovered,with the aim of understanding the formation of grain yield.Eight quantitative trait loci(QTL)controlling spikelet number per panicle and 1000-GW were mapped by sequencing-based genotyping of 150 rice recombinant inbred lines[15].The effects of four QTL from Nipponbare using chromosome segment substitution lines were validated and the QTL were pyramided in rice popular varieties in Asia[15].Yield is a complex trait controlled by many genetic factors associated with yield-component traits[17].Favorable alleles have been “mined”from natural cultivars and wild rice.These rice lines are IR24,Kasalath,Koshihikari,Menghui 63,and Nipponbare,in which functional genes have been identified by association analysis of target traits such as grain weight(GW5)[18],grain size(GS3)[19],grain number(Gn1a)[20],and strong stems and heavy panicles(SCM2/APO1),[21,22].Reasonable combinations of favorable alleles are being used to increase rice yield potential,combining key traits such as excellent plant type,strong stems,and long and heavy panicles with well-filled kernels[16].Alternative pedigree selection methods and use of markers associated with major QTL to target traits can be used by scientists to select high-priority lines for each generation[14].
The objectives of the present study were to(i)investigate the allelic diversity of loci associated with high-yielding parental lines in the varieties NERICA-L-20 and Giza178,with the aim of developing ARS 563 populations,(ii)phenotype and genotype F2,F2:3,F2:4,and F2:5populations using agro-morphological quantitative and qualitative descriptors,yield and yield component traits,and GRiSP polymorphic markers to select new,superior,high-yielding rice lines.
2.Materials and methods
2.1.Agro-morphological measurement and statistical analyses
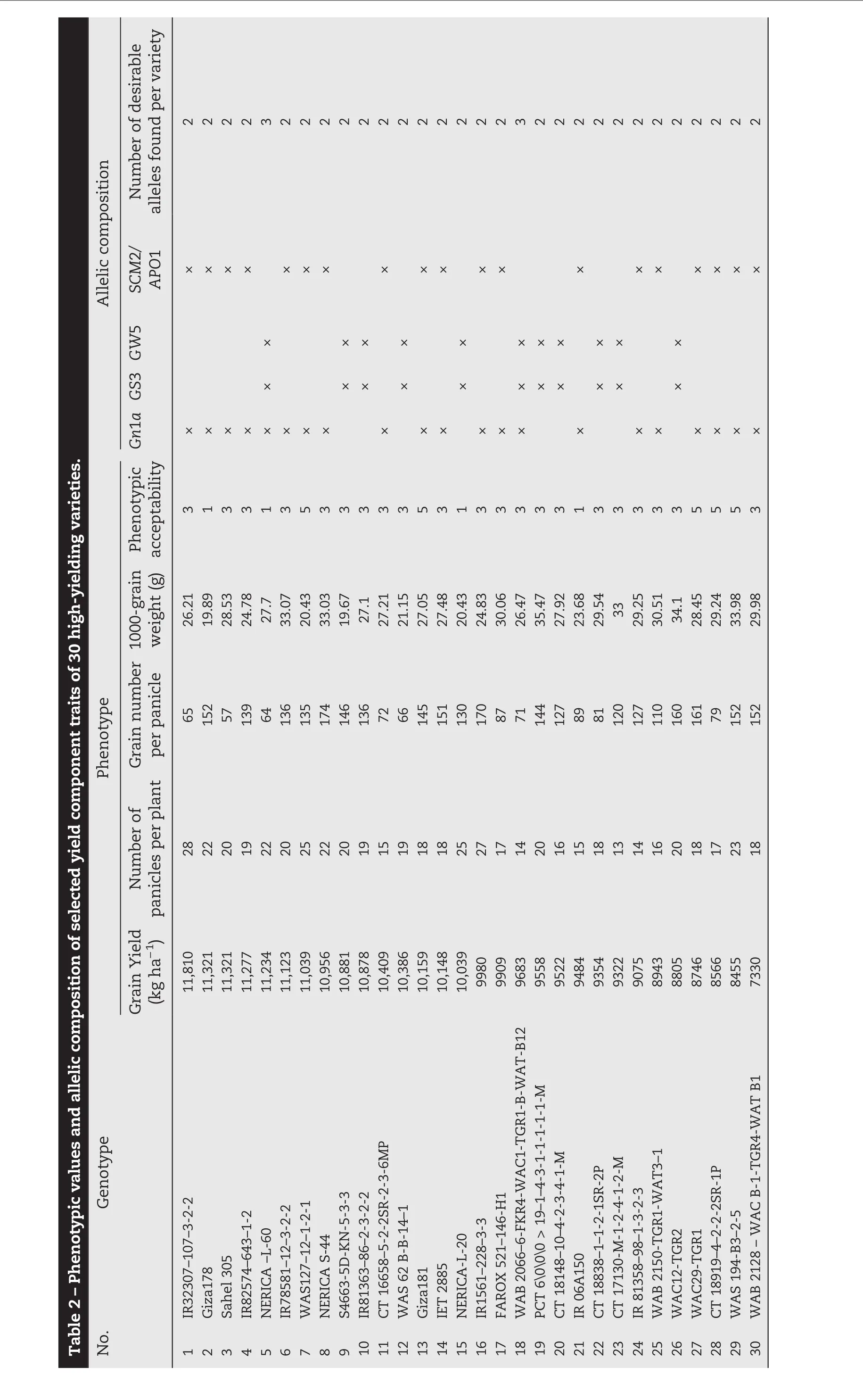
The experiments were conducted at the AfricaRice Regional Research Center in St Louis,Senegal,(16°14′N,16°14′W,9 m a.s.l.).An allelic diversity survey was conducted with 30 high-yielding rice varieties(Fig.1)that were screened and selected from 300 high-yielding indica cultivars from West African countries during the 2012 dry and wet seasons in two locations.Markers polymorphic between NERICA-L-20(AfricaRice)and Giza178(Egypt Research Center)associated with grain weight(GW5,Marker_1 to Marker_3),grain size(GS3,Marker_4 to Marker 6),grain number(Gn1a,Marker_7 to Marker_10)and strong stems and heavy panicles(SCM2/APO1,Marker_11 to Marker_16)were used to show plant performance for yield component traits of each inbred line(Table 1).The F1(ARS 563)progeny derived from crosses between NERICA-L-20 and Giza178 were self-pollinated to generate large F2,F2:3,F2:4,and F2:5populations.Field experiments were conducted twice a year from 2012 to 2014 and F2populations totaling 1000 plants were evaluated during the 2013 dry season.An augmented experimental design laid out in 40 blocks was used to evaluate yield potential.Each block contained two rows of each parent,two checks(Sahel 108 and Sahel 201,released by ISRA Senegal)and 29 F2lines.The parents and checks were replicated in each block.In contrast,a randomized complete block design with three replications was used to evaluate selected F2:3,F2:4,and F2:5lines.The transplanting density was 20 cm between plants within rows and 20 cm between rows.Fertilizers were applied at the rate of 150 kg ha−1as follows:NPK15–15-15at vegetative stage and 60 kg ha−1urea as top dressing at tillering and panicle initiation.Weeds were controlled manually throughout the growing season.The descriptors for rice[23]were used to record total biomass(TB),harvest index(HI),panicle number per square meter(PN/m2),total grain number per panicle(GNP),1000-weight grain(1000-GW),spikelet fertility(SF),and grain yield(GY)for selected F2:3and F2:4.Tiller number at 60 days after planting(T60),plant height at 60 days after planting(H60),and days to heading at 50%flowering(DH50)were added as parameters for selected F2:5plants.Pedigree selection including the two parents and check varieties(Sahel 108 and Sahel 201)was conducted using a phenotypic acceptability parameter rate scaling that ranged from excellent(1)to unacceptable(9)with intermediate values of 3(good),5(fair),and 7(poor).
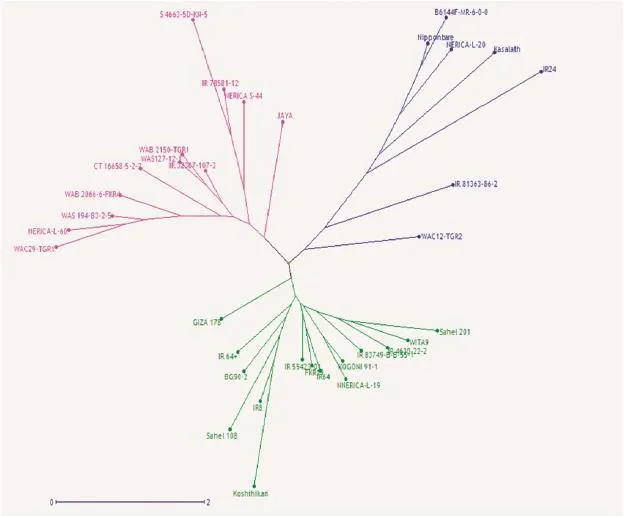
Fig.1–Allelic diversity survey of 30 high-yielding selected varieties and positive checks(IR24,Kasalath,Koshihikari,and Nipponbare)using weighted neighbor-joining clustering of genotype data from 11 polymorphic microsatellite markers associated with major yield-component traits.
ANOVA mixed models were fitted for 10 quantitative traits using XLSTAT software[24].Broad-sense heritability(h2)was calculated using the Breeding Management SystemWorkbench 3.09 software[25]according to the procedure described by[26,27].
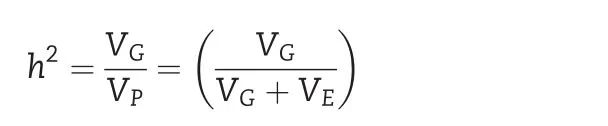
with VG,genotype variance;VP,phenotypic variance;VE,environment variance.
Yield advantage(Yadv)was estimated from grain yield over best parent(GYbp),midparent(GYmidp),and standard check variety(GYsdc)using the method described by[28]:
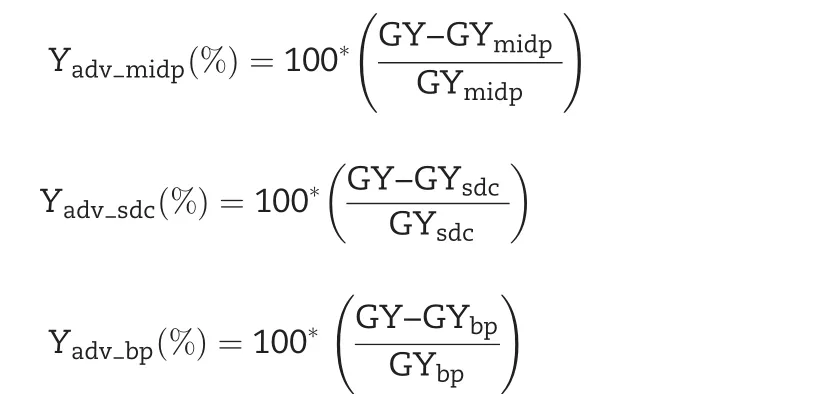
with Yadv_midp(%),yield advantage over the mid-parent;Yadv_sdc(%),yield advantage over the standard check variety;Yadv_bp(%),yield advantage over the best parent;GY,promising line grain yield;GYbp,best-parent grain yield;GYmidp,midparent grain yield;GYsdc,standard check variety grain yield.
2.2.DNA extraction and favorable-allele tracking of 16 SSR and InDel markers associated with major QTL for yield and yield component traits
Genomic DNA was extracted from three-week-old leaves of all selected parental lines,F2,F2:3,and F2:4plants using the CTAB protocol[29]and genotyped with simple sequence repeat(SSR)and InDel GRiSP markers using PCR techniques.Sixteen primers associated with major QTL for yield component traits,as described in Table 1,were used according to the generation.Using the following program,10 μL of each SSR-PCR mixture was amplified:initial denaturation(1 cycle of 94°C for 4 min)followed by 35 amplification cycles including denaturation(94 °C for 1 min);hybridization of primers(55 °C for 1 min),elongation(72 °C for 2 min),and a final elongation(72 °C for 5 min).SSR/InDel-PCR products were separated on 8%polyacrylamide gel with 1x TBE buffer(40 mmol L−1Trizma base-HCl,40 mmol L−1boric acid,and 1 mmol L−1EDTA),stained with 1 μg mL−1bromophenol blue(3XSTR),and visualized with an ultraviolet transilluminator with the image captured by Syngen's G-Box gel imaging system.SSR/InDel(Table 1)profiles were scored and analyzed for allelic similarity(Fig.1)in comparison with Nipponbare,Koshihikari,and IR24 as yield-component positive checks using Darwin software version 6[30].
?
3.Results
3.1.Allelic polymorphic survey with 30 high-yielding varieties
Previously,an allelic polymorphism survey was conducted using the 30 selected high-yielding varieties(Fig.1).Two varieties,NERICA-L-60 and WAB2066–6-FKR4-WAC1-TGR1-B-WATB12,combined three desirable alleles(Gn1a,GS3,and GW5)in their genetic backgrounds,whereas the remaining varieties carried only two favorable alleles,in several allele combinations(Table 2).NERICA-L-20(GS3 and GW5)and Giza178(Gn1a and SCM2/APO1)were used as parental lines to develop ARS 563 populations.A polymorphism survey between the two parental lines was conducted using Nipponbare(GW5),IR24(GS3),and Koshihikari(Gn1a and SCM2/APO1)as positive-allele check varieties to confirm the yield-component trait donor allele coming from each parent.
3.2.Forward breeding in the F2,F2:3,and F2:4 generations
Marked segregation in the F2population was observed for all agronomic traits.A total of 1000 F2plants were phenotyped under field condition and 100 F2:3plants were selected based on their phenotypic acceptability,ranging from 1(excellent)to 3(good)under irrigated growth conditions.These F2:3plants were genotyped using highly polymorphic SSR/InDel markers.
Various numbers of introgressed QTL associated with yield-component traits were found.Forty-four F2:3plants showed two to three introgressions of favorable alleles such as GW5-GS3-SCM2/APO1,GW5-Gn1a-SCM2/APO1,GW5-GS3-Gn1a,Gn1a-GS3-SCM2/APO1,and GW5-GS3-Gn1a-SCM2/APO1 for three favorable allele combinations.However,52 F2:3plants did not show any allele combinations.Four F2:3plants(ARS 563–14,ARS 563–62,ARS 563–286,and ARS 563–41)showed four segments found in chromosomes 1(Gn1a),3(GS3),5(GW5),and 6(SCM2/APO1)and were used for the next marker screening and advance(Fig.2).Usually,the number of selected lines in the next screening could be increased.The stepwise screening method recommended by Sreewongchai et al.[14]was used to select superior,high-yielding new plant types.A total of three F2:5individual plants derived from F2ARS 563–14 and ARS 563–286 families were selected as ideotypes and identified as promising superior high-yielding lines.The alternate phenotype–genotype selection method used to advance progenies from F2to F2:5is described in Fig.3.
3.3.Agro-morphological characterization of selected F2:3 and F2:4 pedigree selection
A total of 53 selected F2:3plants from ARS 563–14,ARS 563–62,ARS 563–286,and ARS 563–41 families were phenotyped and evaluated for high yield potential under field conditions(Table 3).The TB of the F2:3was lowest(1776 g m−2),contrasting with those of the checks Sahel 108(1950 g m−2)and Sahel 201(2106 g m−2),and the two parents.HI was high(0.60)for the F2:3lines and ranged from 0.44 to 0.48 for the two parents.The PN/m2for the F2:3population was 566,exceeding those of both parents,NERICA-L-20(427)and Giza178(515).Moderate(P<0.01)to high phenotypic variation(P<0.0001)was observed for PN/m2,GNP,and HI.GY showed significant(P<0.05)differences,while TB,1000-GW,and SF showed nonsignificant differences.F2:31000-GW was 25.70 g,in contrast to those of the two parents,23.67 and 26.67 g;SF was higher than 75%for the F2:3population and their parents with an average of 76.47%.The average GY of F2:3population was 999 g m−2while the parents showed GY values as follows:NERICA-L-20(921 g m−2)and Giza178(1002 g m−2).Broad-sense heritability(h2)values were high for HI(0.6),PN/m2(0.78),and GNP(0.73)and ranged from moderate to low for other traits.A total of 31 F2:4plants were selected from the 53 selected F2:3plants showing superior high-yielding characteristics,using pedigree selection.The 31 F2:5plants derived from ARS563–14 and ARS563–286 families were used for preliminary yield performance trials.
3.4.Evaluation of selected F2:5 ARS 563–14 and ARS 563–286 lines and preliminary yield performance estimation
The 31 selected plants of the two families ARS 563–14(Table 4)and ARS 563–286(Table 5)including the two parents and two checks were evaluated.Results from 14 F2:5(ARS 563–14)and 17 F2:5(ARS 563–286)showed high phenotypic variation(P<0.0001)for DH50,total grain number per square meter(TGN/m2),panicle length,GY,and SF.However,there were no significant differences for T60,H60,HI,PN/m2,1000-GW,and TB.The mean DH50 was<90 days for the F2:5lines,Sahel 108 and Giza178.The mean values of GNP ranged from 96(ARS 563–286–12–1-4)to 151(ARS 563–14–1-1-1).However,for TGN/m2the values were between 145(ARS 563–14–1-1-1)and 503(ARS 563–286–18-1-1).For 1000-GW,the values obtained were 23.07 g(ARS 563–286–16-1-1)and 28.73 g(ARS 563–286–14–1-1).In addition,the h2values obtained from ten quantitative traits ranged from low (h2<0.2),to moderate(0.2
The three top lines,ARS 563–286–16-1-1,ARS 563–286–5-1-1,and ARS 563–14–10-1-1,showed over 10%yield increase over the values obtained with the best parent,midparent,and standard check variety Sahel 108(Table 6).The 11 best F2:6lines may be inferred to be homozygous for the QTL linked with the yield-component traits.
4.Discussion
?
The ARS563 populations developed from a cross between NERICA-L20 and Giza178 via alternate phenotype–genotype selection combined with pedigree selection could contribute to identifying superior high-yielding rice lines compared with the parents and the standard check.As reported by Khush[8]and Sreewongchai et al.[14],this high yield was due to heterosis resulting from the use of different sources or different genetic backgrounds of the parents.The pedigree selection method is used for selection from segregating populations of crosses in self-pollinated crops and for combination or transgressive breeding.In fact,molecular characterization enabled the identification at an early stage of interesting recombinant lines with common region“introgressed”segments on chromosomes 1(Gn1a),3(GS3),5(GW5),and 6(SMC2/APO1).It also showed that the same segregating line is capable of accumulating varying combinations governing the expression of these different yield component traits[31].The most important way,as reported by Fujita et al.[32],is to understand the enhancement of source size and translocation capacity as well as sink size regarding the phenotypic characteristics of the population.That study showed that near-isogenic lines achieved 13%–36%yield increases with no negative effect on grain appearance.Expression analysis revealed that the gene was expressed in panicles,leaves,roots,and culms supporting the pleiotropic effects on plant architecture.Spikelet number(SPIKE)increased grain yield by 18%in the released indica cultivar Oryza sativa L.and increased the number of spikelets in the genetic background of other popular indica cultivars[32].However,a negative correlation(−0.23)between grain weight and grain number,two major yield component traits,was reported by Venkateswarlu and Visperas[33],depending on lineage source.
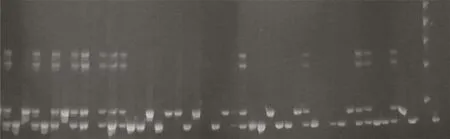
Fig.2–Forward breeding for grain size of selected F2:4lines using RGS1-SSR1(Marker_5).1:Giza178(parent 1);2:NERICA-L-20(parent 2)=IR24=positive check for yield component grain size(GS3);3,4,5,6:F2:4lines genotyped using Marker_5 for grain size(GS3);7:ladder(100 pb).
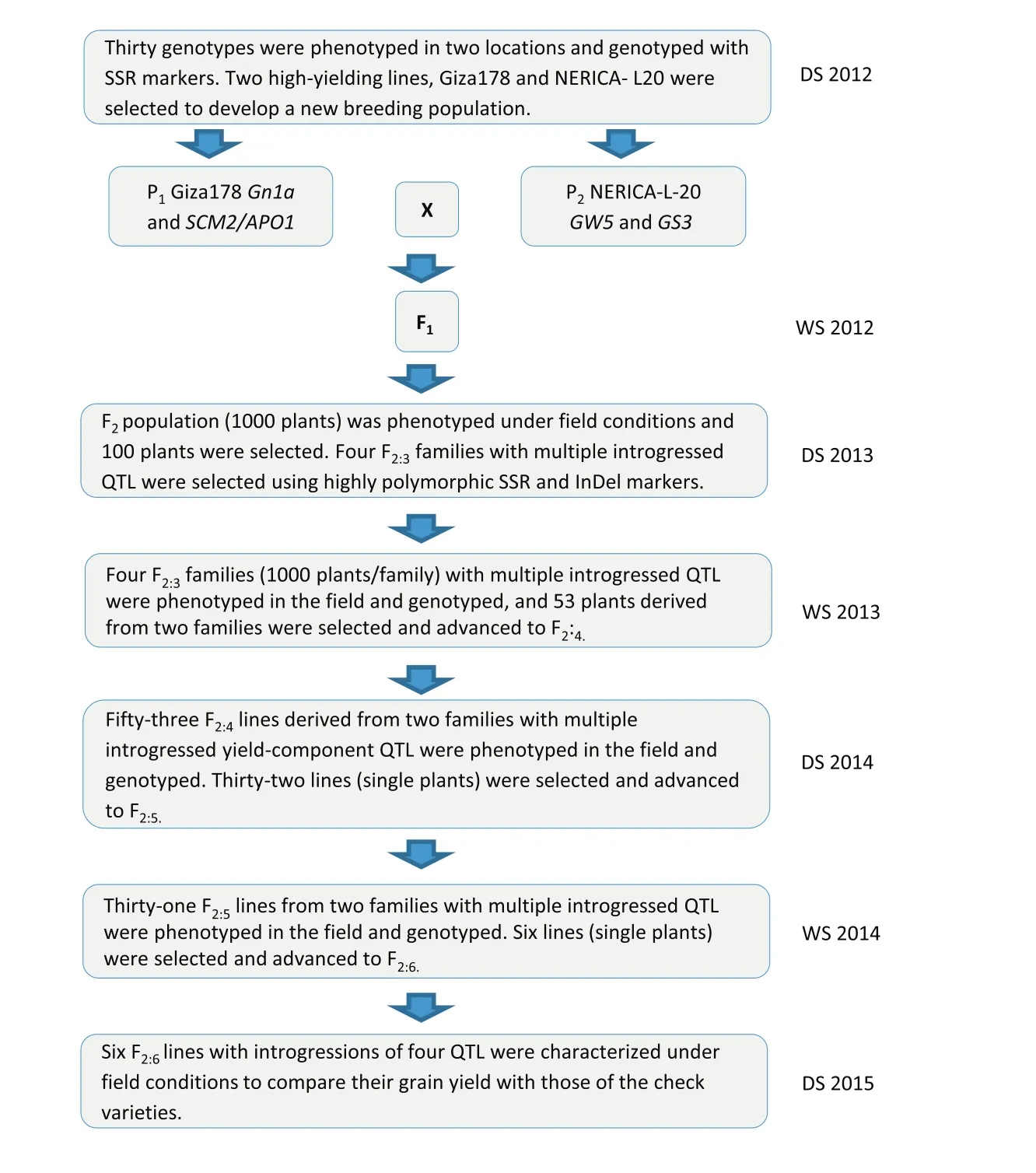
Fig.3–Procedural scheme for advancing selected lines through F2:6generation.
Phenotypic variation was observed in F2:3and F2:5populations with good tillering ability and the semidwarf to intermediate plant height required in irrigated and rainfed lowland growth conditions.On the other hand,F2:5showed strong stems capable of supporting the heavy panicle weight conferred by Giza178(Gn1a and SMC2/APO1).Plant height is one of the main descriptors often used to explain plantarchitecture that supports heavy panicles[34].The selected F2:5lines showed moderate to high heritability for all traits,revealing good to excellent performance of these lines.
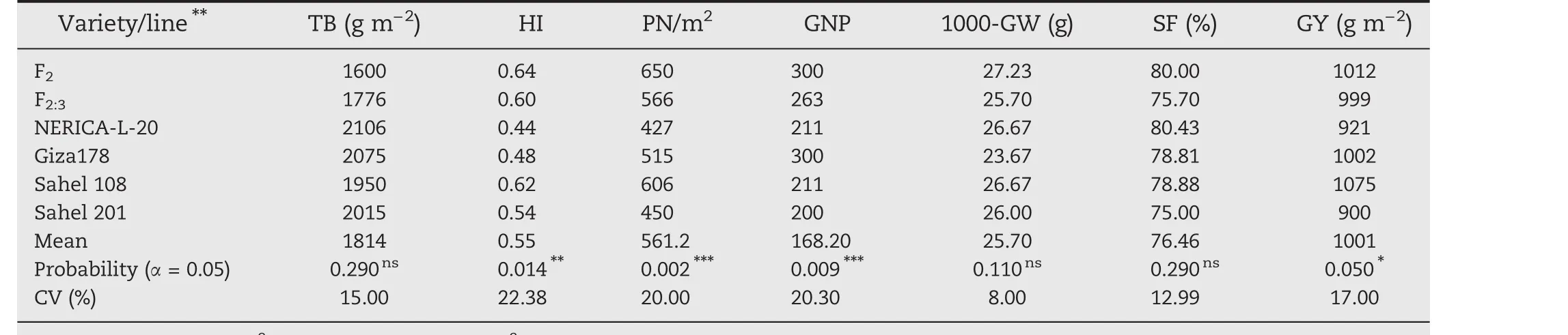
Table 3–Average values of seven traits of the selected lines F2,F2:3 compared with parents and check variegties.

Table 4–Average values of 10 traits of 14 selected F2:5 lines derived from ARS 563–14 compared with parents and check varieties.
The three top selected F2:5lines,ARS 563–286–16-1-1,ARS 563–286–5-1-1,and ARS 563–14–10-1-1,showed an increase of more than 10%grain yield following standard heterosis in comparison with the best check,Sahel 108.
Marker identification of QTL associated with target traits in different crops has contributed to developing methods that combine conventional and molecular breeding to makeprogress in marker-assisted breeding[35].Selection may be applied at any plant growth stage and in small populations.In that case,phenotyping and genotyping by the so-called alternate phenotype–genotype selection method and marker-assisted selection may be used to reduce field trial size by excluding unfavorable genotypes before planting the population in the field[14].Genotype and phenotype are still used to refer to the individual's DNA and traits.The use of markers linked to QTL associated with target traits is contributing to improving the efficiency and precision of conventional plant breeding via marker-assisted selection[36].
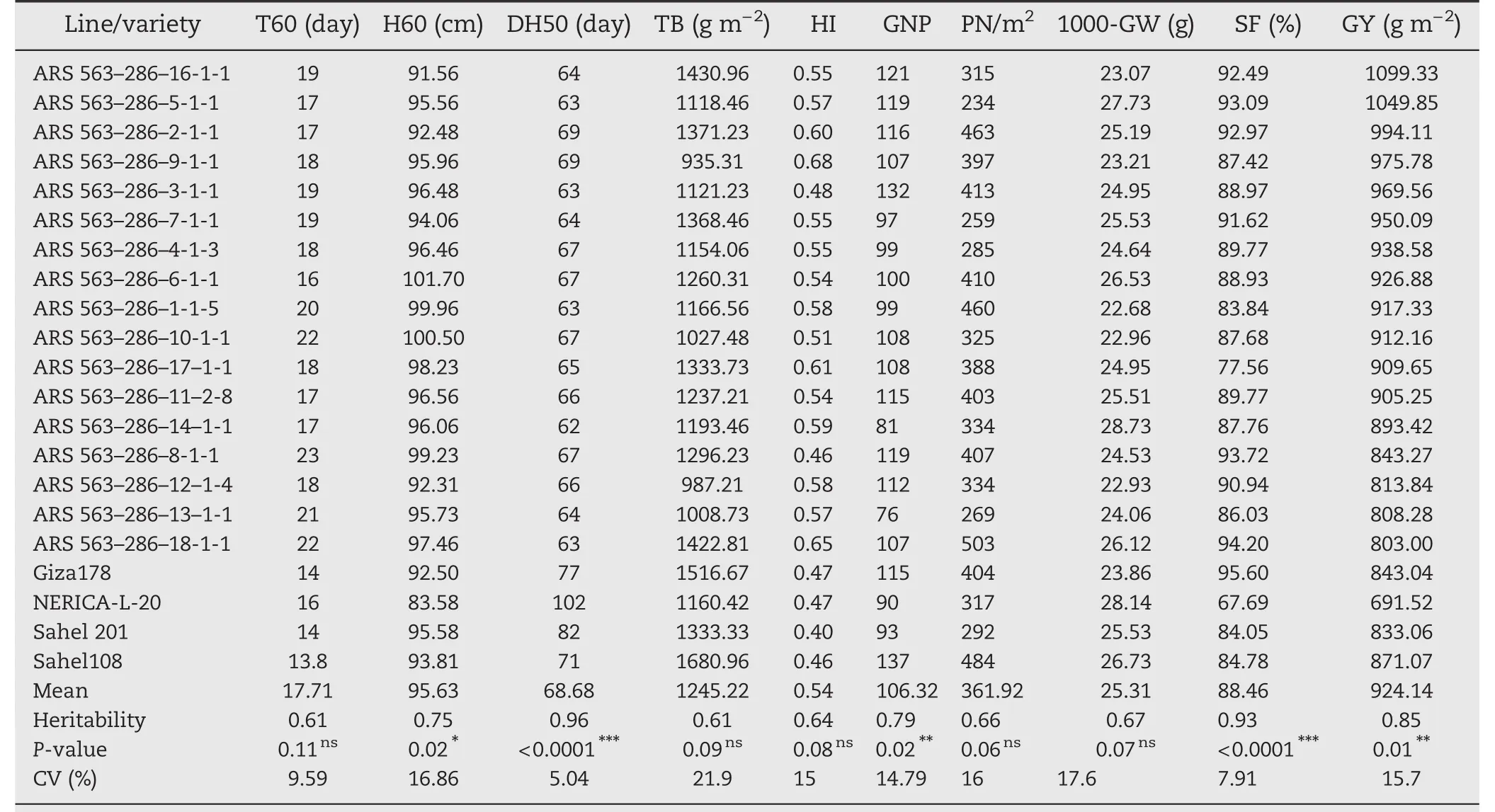
Table 5–Average values of 10 traits of 14 selected F2:5 lines derived from ARS 563–286 compared with parents and check varieties.
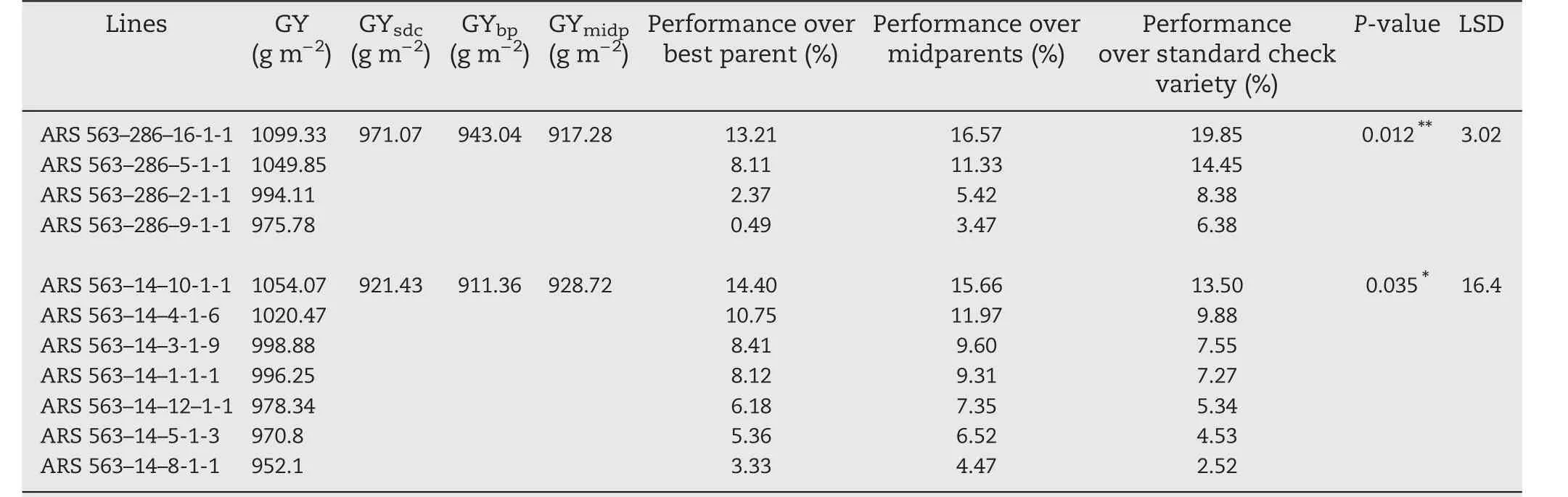
Table 6 –Preliminary yield performance from best selected F2:6 lines derived from ARS 563–286 and ARS 563–14 families.
In conclusion,alternate phenotype-genotype selection may prove useful for accelerating rice breeding programs.
We acknowledge funding to the GRiSP New Frontiers Project(DRPC2012-025).We also thank the irrigated breeding unit and the Biotechnology Laboratory at Africa Rice Saint Louis for assistance provided by the technicians.
[1]Statista,Statistics on “Rice”:world rice acreage from 2008/2009 to 2015/2016(in million hectares),https://www.statista.com/statistics/271969/world-rice-acreage-since-2008/2017.
[2]FAO,Rice Market Monitor(RMM),volume XIX,Issue No.3,October 2016(Rome,Italy,2016).
[3]N.Palmer,Rice roadmap provides an alternative to the quest for “mega-varieties”CIAT Blog http://www.ciatnews.cgiar.org/2011/09/08/rice-roadmap-provides-an-alternative-tothe-quest-for-mega-varieties/2011.
[4]IRRI,Annual Report of the Director General,2005–2006,Volume 16,IRRI,Makati City,Philippines,2006(www.irri.org).
[5]Africa Rice Center(WARDA),Africa Rice Trends:Overview of Recent Developments in the Sub-Saharan Africa Rice Sector,Africa Rice Center Brief,Cotonou,Benin,2007.
[6]Africa Rice Center(AfricaRice),New Breeding Directions at AfricaRice:Beyond NERICA,Africa Rice Center,Cotonou,Benin,2010.
[7]V.O.Sadras,K.G.Cassman,P.Grassini,A.J.Hall,W.G.M.Bastiaanssen,A.G.Laborte,A.E.Milne,G.Sileshi,P.Steduto,Yield Gap Analysis of Field Crops:Methods and Case Studies,FAO Water Reports No.41,Food and Agriculture Organization of the United Nations(FAO)and the Robert B,Daugherty Water for Food Institute at the University of Nebraska(DWFI),Rome,Italy,2015.
[8]G.S.Khush,Breaking the yield frontier of rice,Georgetown Dent.J.35(1995)329–332.
[9]S.B.Peng,G.S.Khush,P.Virk,Q.Y.Tang,Y.B.Zou,Progress in ideotype breeding to increase rice yield potential,Field Crop Res.108(2008)32–38.
[10]P.S.Virk,G.S.Khush,S.B.Peng,Breeding to enhance yield potential of rice at IRRI:the ideotype approach,Int.Rice Res.Notes 29(2004)5–9.
[11]F.M.Xie,B.Hardy,Accelerating Hybrid Rice Development,IRRI,Los Baños,The Philippines,2009.
[12]K.Traore,V.B.Bado,M.K.N'Diaye,Manuel Pratique sur les Normes de Production et de Certification de Semences de Riz,AfricaRice Sahel Station,Saint Louis,Senegal,2012(in French).
[13]T.R.Hargrove,V.L.Cabanilla,W.R.Coffman,Twenty years of rice breeding,Biomed.Sci.38(1988)675–681.
[14]T.Sreewongchai,P.Rattanapol,V.Vichukit,Alternate phenotype-genotype selection method for developing photoperiod intensive,good cooking quality and potential high-yielding rice lines,Kasetsart,J.(Nat.Sci.)48(2014)851–859.
[15]G.Zong,A.H.Wang,L.Wang,G.H.Liang,M.H.Gu,T.Sang,B.Han,A pyramid breeding of eight grain-yield related quantitative trait loci based on marker-assistant and phenotype selection in Rice(Oryza sativa L.),J.Genet.Genomics 39(2012)335–350.
[16]M.Dingkuhn,M.P.Jones,D.E.Johnson,A.Sow,Growth and yield potential of Oryza sativa and O.glaberrima upland rice cultivars and their interspecific progenies,Field Crop Res.57(1998)57–69.
[17]J.Demol,J.P.Baudoin,P.B.Louant,R.Maréchal,G.Mergeai,E.Otoul,L'amélioration des plantes.Application aux principales espèces cultivées en régions tropicales,Les Presses Agronomiques de Gembloux,Gembloux,Belgique,2002(in French).
[18]K.Miura,M.Ikeda,A.Matsubara,X.J.Song,M.Ito,K.Asano,M.Matsuoka,H.Kitano,M.Ashikari,OsSPL14 promotes panicle branching and higher grain productivity in rice,Nat.Genet.42(2010)545–549.
[19]D.W.Xue,Q.Qian,S.Teng,Identification and Utilization of Elite Genes from Elite Germplasms for Yield Improvement,in:W.G.Yan,J.S.Bao(Eds.),Rice-Germplasm,Genetics and Improvement,InTechOpen,Rijeka,Croatia 2014,pp.1–5.
[20]M.Ashikari,H.Sakakibara,S.Y.Lin,T.Yamamoto,T.Takashi,A.Nishimura,E.R.Angeles,Q.Qian,H.Kitano,M.Matsuoka,Cytokinin oxidase regulates rice grain production,Science 309(2005)741–745.
[21]T.Ookawa,H.T.Yano,M.Murata,K.Ando,T.Miura,H.Asano,K.Ochiai,Y.Ikeda,M.Nishitani,R.Ebitani,T.Ozaki,H.Angeles,E.R.Hirasawa,T.M.Matsuoka,New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield,Nat.Commun.1(2010)132.
[22]J.F.A.Griffiths,J.H.Miller,D.T.Suzuki,R.C.Lewontin,W.M.Gelbart,à Introduction,L'analyse Génétique,4th edition de Boeck,Bruxelle,Belgium,2006(in French).
[23]Bioversity International,IRRI and WARDA,Descriptors for Wild and Cultivated Rice(Oryza Spp.),Bioversity International,Rome,Italy,2007(International Rice Research Institute,Los Baños,Philippines;WARDA,Africa Rice Center,Cotonou,Benin).
[24]Addinsoft,XLStat,Version 2015.6,New York,USA,https://www.xlstat.com/en/news/version-2015-6 2015.
[25]Integrated Breeding Platform(IBP),Breeding Management System(BMS),Version,3.0.9,2015,Mexico,D.F.,Mexico,https://www.integratedbreeding.net/.
[26]D.L.Zhao,G.N.Atlin,L.Bastiaans,H.J.Spiertz,Cultivar weed-competitiveness in aerobic rice:heritability,correlated traits,and the potential for indirect selection in weed-free environments,Crop Sci.1(2006)372–380.
[27]H.F.Robinson,R.E.Comstock,P.H.Harvey,Estimates heritability and the degree of dominance in corn,Agron.J.41(1949)353–359.
[28]K.Mather,Biometrical Genetics,the Study of Continuous Variation,2nd ed.Methuen,London,UK,1949.
[29]M.A.Saghai-Maroof,K.M.Soliman,R.A.Jorgensen,R.W.Allard,Ribosomal DNA spacer-length polymorphisms in barley:Mendelian inheritance,chromosomal location,and population dynamics(ribosomal DNA spacer-length variation/restriction fragment-length polymorphisms/Rrnl/Rrn2),Proc.Natl.Acad.Sci.U.S.A.81(1984)8014–8018.
[30]X.Perrier,J.P.Jacquemoud-Collet,DARwin Software,Version 6,http://darwin.cirad.fr/2006.
[31]D.S.Brar,G.S.Khush,Alien introgression in rice,Plant Mol.Biol.35(1997)35–47.
[32]D.Fujita,K.R.Trijatmikoa,A.G.Taglea,M.V.Sapasapa,Y.Koidea,K.Sasakia,N.Tsakirpalogloua,R.B.Gannabana,T.Nishimurad,S.Yanagiharab,Y.Fukutab,T.Koshibad,I.H.Slamet-Loedina,T.Ishimarua,N.Kobayashia,NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars,Proc.Natl.Acad.Sci.U.S.A.110(2013)20431–20436.
[33]B.Venkateswarlu,R.M.Visperas,Source-Sink Relationships in Crop Plants,IRRI,Los Baños,Philippines,1987.
[34]B.P.Caton,A.E.Cope,M.Mortimer,Growth traits of diverse rice cultivars under competition:implications for screening for competitiveness,Field Crop Res.80(2003)157–172.
[35]C.Bertrand,Y.Collard,J.D.Mackill,Marker-assisted selection:an approach for precision plant breeding in the twenty-first century,Philos.Trans.R.Soc.B-Biol.Sci.(2008)557–572.
[36]P.Taylor,R.Lewontin,The Genotype/Phenotype Distinction,in:E.N.Zalta(Ed.),The Stanford Encyclopedia of Philosophy,2017.
杂志排行
The Crop Journal的其它文章
- Integrated physiological and molecular approaches to improvement of abiotic stress tolerance in two pulse crops of the semi-arid tropics
- Genome-wide association study of heat stresstolerance traits in spring-type Brassica napus L.under controlled conditions
- Development and validation of InDel markers for identification of QTL underlying flowering time in soybean
- Zinc partitioning in basmati rice varieties as influenced by Zn fertilization
- Elevated temperature intensity, timing, and duration of exposure affect soybean internode elongation, mainstem node number, and pod number per plant
- Soybean hairy roots produced in vitro by Agrobacterium rhizogenes-mediated transformation
