Gene-based and pathway-based genome-wide association study of alcohol dependence
2015-12-09LingjunZUOClarenceZHANGFrederickSAYWARDKeiHoiCHEUNGKeshengWANGJohnKRYSTALHongyuZHAOXingguangLUO
Lingjun ZUO, Clarence K. ZHANG*, Frederick G. SAYWARD, Kei-Hoi CHEUNG, Kesheng WANG,John H. KRYSTAL, Hongyu ZHAO, Xingguang LUO*
•Original research article•
Gene-based and pathway-based genome-wide association study of alcohol dependence
Lingjun ZUO1, Clarence K. ZHANG2,3*, Frederick G. SAYWARD4,5, Kei-Hoi CHEUNG4, Kesheng WANG6,John H. KRYSTAL1, Hongyu ZHAO2, Xingguang LUO1*
gene-based GWAS; pathway-based GWAS; cell-extracellular matrix interaction pathway;PXN;paxillin; alcohol dependence
1. Introduction
Conventional genome-wide association studies(GWASs) focused on the impact of single nucleotide polymorphisms (SNPs) have identified a large number of significant or suggestive risk genes for alcohol dependence and alcohol consumption.[1-7]However,single-SNP analysis often identifies only a few of the most significant SNPs of the genome and they can only explain a small proportion of the genetic risk for diseases. Accumulating evidence suggests that susceptibility to alcohol dependence emerges from a complex interplay of variants within genes, genomic regions, or gene pathways.[8]Gene variants that individually contribute slightly to alcoholism risk but that may have a more important effect in moderating the impact of other risk genes may be missed by the single-SNP analytic strategy.[9]This problem may be reduced by employing gene-based and pathway-based analytic approaches.
Gene- and pathway-based methods have many advantages over the single-SNP approach.[9,10]First,the functions of many individual SNPs are not wellcharacterized but the functions of whole genes and particular gene pathways are more clearly characterized;many functional studies (e.g., gene expression studies)have been conducted at the gene or pathway level making it possible to assess the association of biological functions with specific genes and pathways. Second,locus heterogeneity (i.e., alleles at different loci cause diseases in different populations) make it difficult to replicate association findings for a single marker, but replication at the gene or pathway level might still be possible when locus heterogeneity exists because a gene or pathway can harbor multiple alleles of the heterogeneous risk markers. Finally, because the numbers of genes and pathways across the genome are much less than the number of single markers, genebased and pathway-based analyses can substantially reduce the number of comparisons considered and,thus, lead to better statistical power.
In the present study, we aimed to identify the risk genes for alcohol dependence and the pathways that are enriched in alcohol dependence-related genes. In view of the fact that the effects of an entire gene that integrates many SNPs would be different from those of a single SNP, and the effects of an entire pathway that integrates many genes would be different from those of a single gene, it is anticipated that the results from gene-based analyses might not be completely consistent with those from pathway-based analyses that use the same dataset, and, similarly, the results from geneand pathway-based analyses might not be completely consistent with those from SNP-based analyses in previous GWASs on the same datasets. In other words,gene- and pathway-based analysis may lead to novel findings.
2. Materials and Methods
2.1 Subjects
The identi fication of the GWAS data used in this analysis is shown in Figure 1. Data from 1409 European-American(EA) cases with alcohol dependence (based on DSM-IV criteria),1518 EA healthy controls,681 African-American(AA) cases, and 508 AA healthy controls were included in this analysis. Detailed demographic data on these subjects were presented in previous GWASs.[11,12]These data came from the merged SAGE (Study of Addiction:Genetics and Environment) and COGA (Collaborative Study On The Genetics of Alcoholism) datasets, which are available on the database of Genotypes and Phenotypes(dbGaP)( https://www.ncbi.nlm.nih.gov/gap). The SAGE dataset (dbGaP access number: phs000092.v1.p1) included COGA, COGEND (Collaborative Genetic Study of Nicotine Dependence), and FSCD (Family Study of Cocaine Dependence) subsets. This COGA subset included in the SAGE dataset was a subset of the main dbGaP COGA dataset (access number:phs000125.v1.p1), so when we merged the SAGE and COGA datasets, one copy of 1477 overlapping subjects were excluded.[11]The projects that collected these data were all approved by the respective institutional review boards, all subjects participating in the projects provided written informed consent, and the current analysis was approved by the institutional review board of Yale University.
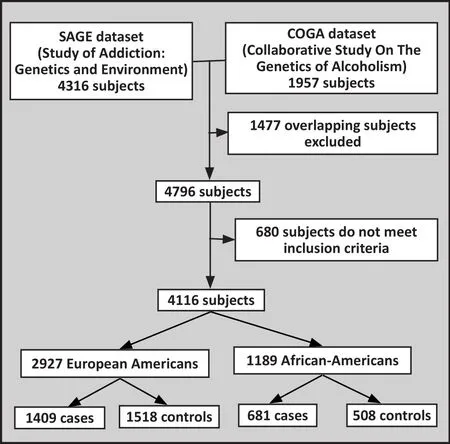
Figure 1. Enrollment of subjects in the study
2.2 Genotyping
All subjects were genotyped on the Illumina Human 1M beadchip. Phenotype and genotype data were rigorously cleaned before association analysis. Subjects with poor genotypic data, allele discordance, problematic sample identification (relatedness, misidentification,misspecification), duplicated identifiers, gender or chromosomal anomalies, ethnicity issues (including missing information, non-EA or AA, mismatch between self- and genetically-inferred ethnicity), or with a missing genotype call rate ≥2% across all SNPs were excluded. Furthermore, SNPs with allele discordance,chromosomal anomalies or batch effect, SNPs with an overall missing genotype call rate ≥2%, monomorphic SNPs, SNPs with minor allele frequencies <0.01 in either EAs or AAs, and SNPs that deviated from Hardy-Weinberg equilibrium (p<10−4) within EA or AA controls were also excluded. This selection process yielded 805,814 SNPs in EAs and 895,714 SNPs in AAs.[11,12]
2.3 Statistical methods
The genotyping data on autosomes were extracted from an Oracle database and stored efficiently in flat files for gene- and pathway-based analysis.
2.3.1 Gene-based GWAS analysis
The genotype was con figured into a genotype score of 1, 2, or 3: 1 represented a minor allele homozygote, 2 represented a heterozygote, and 3 represented a major allele homozygote. SNPs were mapped to known genes/exons/introns boundaries obtained from the National Center for Biotechnology Information (NCBI). Principal component analysis was applied to the SNPs within the defined gene boundary and then the components which explained at least 85% of the variation were used as explanatory variables in the regression to explain disease status. The disease status was de fined as 2 for alcohol dependence and 1 for healthy control. The gene level score was defined as the p-value for the genebased association from this multiple regression.
Gene flanking is defined as increasing the SNPs associated with a gene by extending the gene region by a number of bases in the 5’ and 3’ directions. By doing this, SNPs that may be involved in the transcription process are considered in the analysis. In the discovery analysis in EAs, 50Kb flanking regions were chosen. The 10Kb flanking regions were also explored for top-ranked genes. The top-ranked risk genes identi fied in EAs were also replicated in AAs (with 50Kb flanking regions).
2.3.2 Pathway-based GWAS analysis
Pathway annotation was obtained from the collection of pathways curated by the Molecular Signatures database(MSigDB) using seven public databases: BioCarta, Gene arrays, BioSciences Corp, KEGG, REACTOME, Sigma-Aldrich pathways, Signal transduction knowledge environment, and Signaling gateway (http://www.broadinstitute.org/gsea/msigdb/collection_details.jsp#CP). The gene set enrichment method was used to determine the pathway enrichment.[13]The test statistic was calculated as the negative sum of the log p-values for each gene assigned to the pathway. The enrichment was determined by randomly permuting the gene scores (5000 times) and recalculating the test statistic for each pathway. The p-value of each pathway was the percentage of the permuted test statistics larger than the observed p-value. The top-ranked pathways identified in EAs were replicated in AAs (with 50Kb flanking regions).
2.3.3 Correction for multiple testing in gene- and pathway-based GWAS analyses
A total of 26 307 genes and 221 pathways were analyzed. The significance levels (α) for gene- and pathway-based GWAS tests were corrected by the Bonferroni correction and, thus, set at 1.9E6 and 2.3E4,respectively. P-values larger than α but less than 0.05 were labelled as ‘nominally signi ficant’.
3. Results
A total of 2464 genes were nominally associated with alcohol dependence in EAs (p<0.05). The 20 top-ranked risk genes (based on the level of statistical signi ficance)are listed in Table 1. After correction for multiple testing (α=1.9E-6), the paxillin gene (PXN) (±50kb)was significantly associated with alcohol dependence(p=3.9E-7). If flanking regions were reduced to ±10kb,PXN(±10kb) remained significantly associated with alcohol dependence in EAs (p<E-8), and the other 19 top-ranked risk genes remained nominally significant(p<0.05).

Table 1. Top-ranked and replicable genes for alcohol dependence
Among the 2464 nominally associated genes in EA,129 were nominally replicable in AAs (p<0.05) (data not shown). As shown in Table 1, six of these genes (ZNF256,CPLX2, LOC646820, SLC38A1, PGBD3, andAP3S2) were associated with alcohol dependence at thep<0.01 level in both EAs and AAs. Only one of these six genes,SLC38A1, is a component of a nominally significant pathway (the ‘amino acid transport across the plasma membrane’ pathway, pathway #18 in Table 2). Among the other 123 nominally replicable genes, only two genes are components of top-ranked pathways:BADbelongs to the ‘VEGF signaling’ pathway (pathway #4 in Table 2) andIQSEC3belongs to the ‘endocytosis’pathway (pathway #6 in Table 2).
Twenty pathways enriched in alcohol dependencerelated genes in EAs are listed in Table 2, including the 17 top-ranked pathways (based on the level of statistical signi ficance) and 3 other important pathways; pathway#18 (the ‘amino acid transport across the plasma membrane’ pathway) is the only nominally significant pathway that contains one of the six replicable genes with p-values <0.01 shown in Table 1 (SLC38A1),pathway #19 was previously reported to be related to addiction, and pathway #20 was nominally replicable in both EA and AA. Using 50kb flanking regions in the analysis of EAs, the top-ranked (#1) risk pathway was the‘cell-extracellular matrix interactions’ pathway (RSU1,LIMS1, LIMS2, ARHGEF6, FERMT2, ACTN1, BLIM1, FLNC,ITGB1, PXN, FLNA, VASP, ILK, TESK1, PARVB,andPARVA)(p<2.0E-4). Two other pathways of particular interest were the ‘VEGF signaling’ pathway (#4) (PXN, BAD,HRAS, NRAS, et al.) (p=1.4E-3) because it contains the nominally replicableBADgene and the ‘endocytosis’pathway (#6) (IQSEC3, HRAS, et al.) (p=7.4E-3) because it contains the nominally replicableIQSEC3gene.
After correction for multiple testing, the only pathway that remained significantly associated with alcohol dependence (p<2.3E-4) was pathway #1. If 10kb flanking regions were set, the association of all of the listed pathways with alcohol dependence in EAs remained nominally signi ficant (p<0.05), but none of them were statistically significant after correction for multiple testing. The two pathways most strongly associated with alcohol dependence when using 10kb flanking regions were the ‘Na+/Cl- dependent neurotransmitter transporters’ pathway (#15) (SLC6A1,SLC6A2, SLC6A3, SLC6A5, SLC6A6, SLC6A7, SLC6A9,SLC6A11, SLC6A12, SLC6A13, SLC6A14, SLC6A15,SLC6A18, SLC6A19, SLC6A20, SLC18A1, SLC18A2, andSLC22A2) and the ‘amino acid transport across the plasma membrane’ pathway (#18) (SLC1A4, SLC1A5,SLC3A1, SLC3A2, SLC6A6, SLC6A12, SLC6A14, SLC6A15,SLC6A18, SLC6A19, SLC6A20, SLC7A1, SLC7A2, SLC7A3,SLC7A5, SLC7A6, SLC7A7, SLC7A8, SLC7A9, SLC7A10,SLC7A11, SLC16A10, SLC36A1, SLC36A2, SLC38A1,SLC38A2, SLC38A3, SLC38A4, SLC38A5, SLC43A1, andSLC43A2) (both p=1.8E-3).
As shown in Table 2, there were 2 nominally replicable pathways (based on 50kb flanking) enriched in alcohol dependence-related genes in both EAs(0.015≤p≤0.035) and AAs (0.025≤p≤0.050): the‘Na+/Cl- dependent neurotransmitter transporters’pathway (#15) (speci fied above), and the ‘other glycan degradation’ pathway (#20) (AGA, HEXA, HEXB, ENGASE,FUCA2, FUCA1, MANBA, GLB1, MAN2C1, MAN2B2,NEU1, NEU3, MAN2B1, NEU2, GBA, andNEU4).
4. Discussion
4.1 Main findings
In the present study, we found significant genomewide replicable risk genes and risk pathways that were associated with alcohol dependence. Incorporating the biological, bioinformatic, statistical, and association evidence with previous reports of these genes and pathways, the ‘cell-extracellular matrix interactions’pathway (#1) and thePXNgene (which encodes paxillin) were the most promising risk factors for alcohol dependence; their association with alcohol dependence remained statistically significant after adjusting for multiple testing using the Bonferroni correction.
The ‘cell-extracellular matrix (ECM) interactions’pathway plays a critical role in regulating a variety of cellular processes in multi-cellular organisms including motility, shape change, survival, proliferation, and differentiation. Cell-ECM contact is mediated by transmembrane cell adhesion receptors (integrins)that interact with extracellular matrix proteins and cytoplasmic adaptor proteins. Many of these adaptor proteins physically interact with the actin cytoskeleton or function in signal transduction.[14]Paxillin is an important component of this pathway that binds directly to α-integrins.
ThePXNgene was significantly associated with alcohol dependence in the present study, suggesting the possible role of paxillin in alcoholism. Paxillin is expressed in multiple tissues (including the brain) where it acts as a multidomain scaffolding protein for bringing together signaling molecules, structural components,and regulatory proteins that control the adhesion and organization of the internal cytoskeleton for processes such as cell migration (reviewed in[15]).
Paxillin is also a component of the ‘VEGF signaling’pathway (#4). This pathway is enriched in alcohol dependence-related genes in EAs, though the association(p=1.4E-3) does not reach our criteria for statistical significance. This pathway has been implicated in stress reactivity and in the symptoms of mood disorders,[16]potential contributors to the risk for alcohol dependence.[17]It has also been associated with drug addiction (including alcoholism) (p=3.2E-3)in a previous report.[18]Interestingly, theBAD(BCL2-associated agonist of cell death) gene also belongs to this pathway; we found a strong, but not statistically significant, association ofBADto alcohol dependence both in EAs and AAs, supporting the possible role of the‘VEGF signaling’ pathway in alcohol dependence.
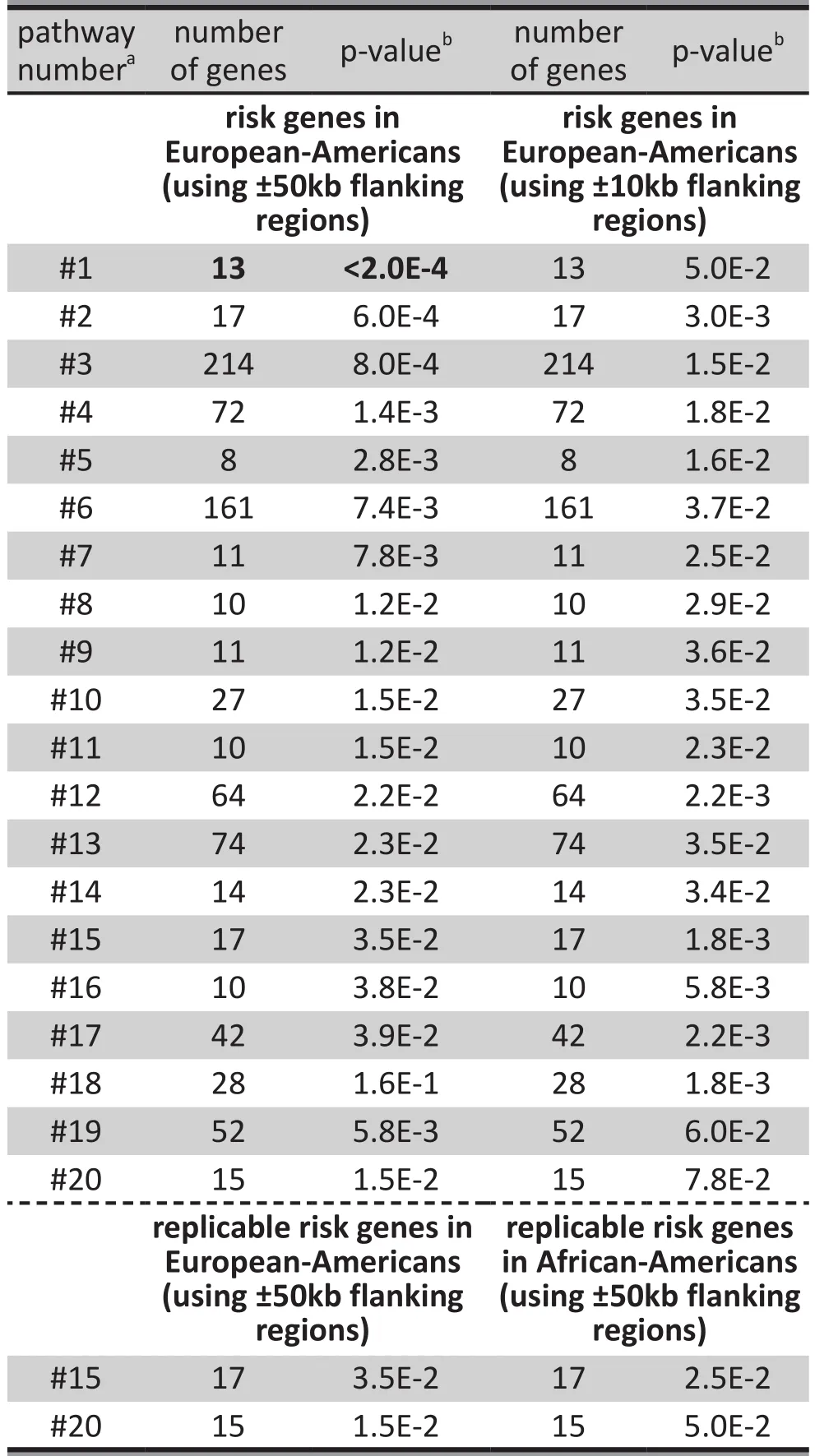
Table 2. Top-ranked and replicable risk pathways for alcohol dependence
Pathways comprehensively integrate information from multiple genes. The complexity of pathway structure makes the replicability of pathway-wise associations very difficult. Replications between homogeneous samples may be relatively common,but replications between genetically heterogeneous samples, such as that between EAs and AAs, would be relatively uncommon. Therefore, replications of pathway-disease associations between EAs and AAs may indicate a functional relationship between the specific pathways and the disease of interest. We identified two replicable pathways for associations with alcohol dependence across EAs and AAs: the‘Na+/Cl- dependent neurotransmitter transporters’pathway (pathway #15 in Table 2) and the ‘other glycan degradation’ pathway (pathway #20 in Table 2). Among all pathways we studied, pathway #15 had the strongest association with alcohol dependence when 10Kb flanking regions were set (p=1.8E-3). All the genes within this pathway are neurotransmitter transporter genes, encoding proteins that mediate neurotransmitter uptake and, thus, terminate a synaptic signal. These transporters are mainly present in the central and peripheral nervous systems[19]where they mediate transport of GABA (gamma-aminobutyric acid),norepinephrine, dopamine, serotonin, glycine, taurine,L-proline, creatine, and betaine. These genes have been associated with several neuropsychiatric conditions;for example,SLC6A3(the dopamine transporter gene,DAT1) andSLC18A2(the monoamine transporter gene)have been associated with alcohol dependence[20-26]and smoking.[27-31]
Another pathway of interest is pathway #18 (the‘amino acid transport across the plasma membrane’pathway) that had a non-significant enrichment of alcohol dependence-related genes in EAs. This pathway was not replicable in AAs, but it contained an important gene,SLC38A1, that was replicable in both the EA population (p=5.7E-3) and the AA population (p=9.9E-3).All genes within this pathway belong to the solute carrier (SLC) family, including amino acid transporter genes which encode proteins that transport amino acid across plasma membranes. These proteins are critical to the uptake of amino acids from the gut, from the renal proximal tubules, and in cells throughout the body where amino acids are required for neurotransmission and for the synthesis of proteins and metabolic intermediates.[32]This pathway is a component of the 18 systems identi fied in physiological studies that mediate amino acid transport, each characterized by its amino acid substrates, its pH sensitivity, and its association(or not) with ion transport.[33]TheSLC38A1(amino acid transporter A1) gene within pathway #18 plays an essential role in the uptake of nutrients, production of energy, chemical metabolism, detoxification, and neurotransmitter cycling. It is an important transporter of glutamine – an intermediate in the detoxification of ammonia and in the production of urea. Glutamine serves as a precursor for the synaptic transmitters glutamate and GABA, both of which have been implicated in the neurobiology of alcohol intoxication and withdrawal.[34]Moreover, glutamate and GABA signaling pathways have been associated with alcohol dependence in a recent pathway-based association study.[35]
Several other top-ranked pathways identi fied in EAs in our study have also been identi fied as potential risk factors for drug addiction and alcoholism in previous reports.[18]These include the ‘long term depression (LTD)’pathway (#12) (p=2.2E-2 in our study, and p=2.1E-7 in a previous study[18]); the ‘Fc epsilon RI signaling’ pathway(#13) (p=2.3E-2 in our study, and p=6.9E-3 in a previous study[18]); and the ‘amyotrophic lateral sclerosis’pathway (#19) (p=5.8E-3 in our study, and p=3.9E-5 in a previous study[18]). Cerebellar LTD is thought to be a molecular and cellular basis for cerebellar learning which promotes the type of neuroplasticity that underlies development and recovery from addiction; a hypothesis that is supported by the finding that many molecular substrates of addiction are shared with other forms of learning.[36,37]Moreover, the LTD pathway has also been found to be enriched in genes associated with smoking cessation, a close phenotype to alcohol dependence.[38]The ‘Fc epsilon RI signaling’ pathway (#13) in mast cells is initiated by the interaction of an antigen (Ag) with IgE which is bound to the extracellular component of the alpha chain of Fc epsilon RI; the activated pathway is regulated both positively and negatively by the interactions of numerous signaling molecules. Activated mast cells release preformed granules containing biogenic amines, especially histamines—the chemicals that regulate alcohol-related behaviors in the brain.[39,40]The ‘amyotrophic lateral sclerosis (ALS)’ pathway (#19)may be involved in glutamate dysregulation, oxidative stress, and mitochondrial damage which may, in turn, be associated with the development of alcohol dependence[34]and alcohol-related neurotoxicity.[41]
4.2 Limitations
With the exception of a significant association of the PXN gene and the ‘cell-extracellular matrix interactions’ pathway in EAs, none of the other topranked risk genes or risk pathways identified in the present study remained significantly associated with alcohol dependence after the results were adjusted for multiple testing using the Bonferroni correction. Further replication studies with even larger samples will be needed to con firm or disprove their relevance to alcohol dependence.
4.3 Implications
In summary, a gene- and pathway-based reanalysis of prior GWAS data provides new evidence highlighting several genes and biological signaling processes that may be related to the risk for alcohol dependence. These pathways converge on glutamate neurotransmission, a process previously implicated in both the neurobiology and treatment of alcoholism. These findings may be helpful in linking genes implicated in the heritable risk for alcohol dependence to this underlying neurobiology.
Acknowledgments
We thank the NIH GWAS Data Repository, the contributing investigator who provided the phenotype and genotype data from their original studies, and the primary funding organization that supported the contributing study. The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gap through dbGaP accession numbers phs000092.v1.p1 and phs000125.v1.p1.
Con flict of interest
None of the authors report any conflict of interest related to this manuscript.
Funding
This work was supported in part by National Institute on Drug Abuse (NIDA) grants K01 DA029643 and R01DA016750, National Institute on Alcohol Abuse and Alcoholism (NIAAA) grants R21 AA021380 and R21 AA020319, the National Alliance for Research on Schizophrenia and Depression (NARSAD) Award 17616 (L.Z.) and ABMRF/The Foundation for Alcohol Research (L.Z.). Funding and other supports for phenotype and genotype data were provided through the National Institutes of Health (NIH) Genes,Environment and Health Initiative (GEI) (U01HG004422,U01HG004436 and U01HG004438); the GENEVA Coordinating Center (U01HG004446); the NIAAA(U10AA008401, R01AA013320, P60AA011998); the NIDA (R01DA013423); the National Cancer Institute(P01 CA089392); the NIH contract ‘High throughput genotyping for studying the genetic contributions to human disease’ (HHSN268200782096C); the Center for Inherited Disease Research (CIDR); and the National Center for Biotechnology Information. Genotyping was performed at the Johns Hopkins University Center for Inherited Disease Research.
Ethics approval
The protocols described in the paper were all approved by the relevant institutional review boards. All subjects were de-identified in this study and the study was approved by the institutional review board at Yale University.
Informed consent
All subjects provided written informed consent to participate in the projects at each of the participating institutions.
1. Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M,Zill P, et al. Genome-wide association study of alcohol dependence.Arch Gen Psychiatry. 2009; 66(7): 773-784. doi:http://dx.doi.org/10.1001/archgenpsychiatry.2009.83
2. Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C,Pugh E, et al. A genome-wide association study of alcohol dependence.Proc Natl Acad Sci U S A. 2010; 107(11): 5082-5087. doi: http://dx.doi.org/10.1073/pnas.0911109107
3. Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN,Almasy L, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11.Alcohol Clin Exp Res. 2010; 34(5): 840-852. doi: http://dx.doi.org/10.1111/j.1530-0277.2010.01156.x
4. Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM,Lind PA, et al. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications.Biological Psychiatry. 2011; 70(6): 513-518. doi:http://dx.doi.org/10.1016/j.biopsych.2011.02.028
5. Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption.Proc Natl Acad Sci U S A. 2011; 108(17): 7119-7124. doi: http://dx.doi.org/10.1073/pnas.1017288108
6. Zuo L, Lu L, Tan Y, Pan X, Cai Y, Wang X, et al. Genome-wide association discoveries of alcohol dependence.Am J Addict.2014; 23: 526-539. doi: http://dx.doi.org/10.1111/j.1521-0391.2014.12147.x
7. Zuo L, Wang K, Zhang X, J.H. K, Li CR, Zhang F, et al. NKAIN1-SERINC2 is a functional, replicable and genome-wide significant risk region specific for alcohol dependence in subjects of European descent.Drug Alcohol Depend.2013; 129: 254-264. doi: http://dx.doi.org/10.1016/j.drugalcdep.2013.02.006
8. Gui H, Li M, Sham PC, Cherny SS. Comparisons of seven algorithms for pathway analysis using the WTCCC Crohn’s Disease dataset.BMC Res Notes. 2011; 4: 386. doi: http://dx.doi.org/10.1186/1756-0500-4-386
9. Peng G, Luo L, Siu H, Zhu Y, Hu P, Hong S, et al. Gene and pathway-based second-wave analysis of genome-wide association studies.Eur J Hum Genet. 2010; 18(1): 111-117.doi: http://dx.doi.org/10.1038/ejhg.2009.115
10. Zamar D, Tripp B, Ellis G, Daley D. Path: a tool to facilitate pathway-based genetic association analysis.Bioinformatics.2009; 25(18): 2444-2446. doi: http://dx.doi.org/10.1093/bioinformatics/btp431
11. Zuo L, Gelernter J, Zhang CK, Zhao H, Lu L, Kranzler HR, et al. Genome-wide association study of alcohol dependence implicates KIAA0040 on chromosome 1q.Neuropsychopharmacology. 2012; 37(2): 557-566. doi:http://dx.doi.org/10.1038/npp.2011.229
12. Zuo L, Zhang CK, Wang F, Li CS, Zhao H, Lu L, et al. A novel,functional and replicable risk gene region for alcohol dependence identified by genome-wide association study.PLoS One. 2011; 6(11): e26726. doi: http://dx.doi.org/10.1371/journal.pone.0026726
13. Ballard D, Abraham C, Cho J, Zhao H. Pathway analysis comparison using Crohn’s disease genome wide association studies.BMC Med Genomics. 2010; 3: 25
14. Sepulveda JL, Gkretsi V, Wu C. Assembly and signaling of adhesion complexes.Curr Top Dev Biol. 2005; 68: 183-225.doi: http://dx.doi.org/10.1016/S0070-2153(05)68007-6
15. Deakin NO, Turner CE. Paxillin comes of age. J Cell Sci. 2008;121(Pt 15): 2435-2444. doi: http://dx.doi.org/10.1242/jcs.018044
16. Newton SS, Duman RS. Regulation of neurogenesis and angiogenesis in depression.Curr Neurovasc Res. 2004; 1(3):261-267. doi: http://dx.doi.org/10.2174/1567202043362388
17. Sinha R, Li CS. Imaging stress- and cue-induced drug and alcohol craving: association with relapse and clinical implications.Drug Alcohol Rev. 2007; 26(1): 25-31 18. Li CY, Mao X, Wei L. Genes and (common) pathways underlying drug addiction.PLoS Comput Biol. 2008; 4(1): e2.doi: http://dx.doi.org/10.1371/journal.pcbi.0040002
19. Chen NH, Reith ME, Quick MW. Synaptic uptake and beyond:the sodium- and chloride-dependent neurotransmitter transporter family SLC6.Pflugers Arch. 2004; 447(5): 519-531
20. Lind PA, Eriksson CJ, Wilhelmsen KC. Association between harmful alcohol consumption behavior and dopamine transporter (DAT1) gene polymorphisms in a male Finnish population.Psychiatr Genet. 2009; 19(3): 117-125. doi:http://dx.doi.org/10.1097/YPG.0b013e32832a4f7b
21. Wernicke C, Smolka M, Gallinat J, Winterer G, Schmidt LG, Rommelspacher H. Evidence for the importance of the human dopamine transporter gene for withdrawal symptomatology of alcoholics in a German population.Neurosci Lett. 2002; 333(1): 45-48
22. Gorwood P, Limosin F, Batel P, Hamon M, Ades J, Boni C. The A9 allele of the dopamine transporter gene is associated with delirium tremens and alcohol-withdrawal seizure.Biol Psychiatry. 2003; 53(1): 85-92
23. Ueno S, Nakamura M, Mikami M, Kondoh K, Ishiguro H,Arinami T, et al. Identification of a novel polymorphism of the human dopamine transporter (DAT1) gene and the signi ficant association with alcoholism.Mol Psychiatry. 1999;4(6): 552-557
24. Vaske J, Beaver KM, Wright JP, Boisvert D, Schnupp R. An interaction between DAT1 and having an alcoholic father predicts serious alcohol problems in a sample of males.Drug Alcohol Depend. 2009; 104(1-2): 17-22. doi: http://dx.doi.org/10.1016/j.drugalcdep.2009.01.020
25. Kohnke MD, Batra A, Kolb W, Kohnke AM, Lutz U, Schick S,et al. Association of the dopamine transporter gene with alcoholism.Alcohol Alcohol.2005; 40(5): 339-342
26. Schwab SG, Franke PE, Hoefgen B, Guttenthaler V,Lichtermann D, Trixler M, et al. Association of DNA polymorphisms in the synaptic vesicular amine transporter gene (SLC18A2) with alcohol and nicotine dependence.Neuropsychopharmacology.2005; 30(12): 2263-2268
27. Ling D, Niu T, Feng Y, Xing H, Xu X. Association between polymorphism of the dopamine transporter gene and early smoking onset: an interaction risk on nicotine dependence.J Hum Genet. 2004; 49(1): 35-39
28. Sieminska A, Buczkowski K, Jassem E, Niedoszytko M,Tkacz E. Influences of polymorphic variants of DRD2 and SLC6A3 genes, and their combinations on smoking in Polish population.BMC Med Genet. 2009; 10: 92. doi: http://dx.doi.org/10.1186/1471-2350-10-92
29. Timberlake DS, Haberstick BC, Lessem JM, Smolen A,Ehringer M, Hewitt JK, et al. An association between the DAT1 polymorphism and smoking behavior in young adults from the National Longitudinal Study of Adolescent Health.Health Psychol. 2006; 25(2): 190-197
30. Stapleton JA, Sutherland G, O’Gara C. Association between dopamine transporter genotypes and smoking cessation: a meta-analysis.Addict Biol. 2007; 12(2): 221-226. doi: http://dx.doi.org/10.1111/j.1369-1600.2007.00058.x
31. Uhl GR, Drgon T, Johnson C, Walther D, David SP, Aveyard P, et al. Genome-wide association for smoking cessation success: participants in the Patch in Practice trial of nicotine replacement.Pharmacogenomics. 2010; 11(3): 357-367. doi:http://dx.doi.org/10.2217/pgs.09.156
32. Yu N, Seo J, Rho K, Jang Y, Park J, Kim WK, et al. hiPathDB:a human-integrated pathway database with facile visualization.Nucleic Acids Res. 2012; 40(Database issue):D797-802. doi: http://dx.doi.org/10.1093/nar/gkr1127
33. Broer S. Amino acid transport across mammalian intestinal and renal epithelia.Physiol Rev. 2008; 88(1): 249-286. doi:http://dx.doi.org/10.1152/physrev.00018.2006
34. Krystal JH, Petrakis IL, Mason G, Trevisan L, D’Souza DC.N-methyl-D-aspartate glutamate receptors and alcoholism:reward, dependence, treatment, and vulnerability.Pharmacol Ther. 2003; 99(1): 79-94
35. Reimers MA, Riley BP, Kalsi G, Kertes DA, Kendler KS.Pathway based analysis of genotypes in relation to alcohol dependence.Pharmacogenomics J. 2011; 12(4): 342-348.doi: http://dx.doi.org/10.1038/tpj.2011.1010
36. Knackstedt LA, Moussawi K, Lalumiere R, Schwendt M,Klugmann M, Kalivas PW. Extinction training after cocaine self-administration induces glutamatergic plasticity to inhibit cocaine seeking.J Neurosci. 2010; 30(23): 7984-7992. doi:http://dx.doi.org/10.1523/JNEUROSCI.1244-10.2010
37. Nestler EJ. Common molecular and cellular substrates of addiction and memory.Neurobiol Learn Mem. 2002; 78(3):637-647
38. Wang J, Li MD. Common and unique biological pathways associated with smoking initiation/progression,nicotine dependence, and smoking cessation.Neuropsychopharmacology. 2010; 35(3): 702-719. doi:http://dx.doi.org/10.1038/npp.2009.178
39. Zimatkin SM, Anichtchik OV. Alcohol-histamine interactions.Alcohol Alcohol. 1999; 34(2): 141-147. doi: http://dx.doi.org/10.1093/alcalc/34.2.141
40. Panula P, Nuutinen S. Histamine and H3 receptor in alcoholrelated behaviors.J Pharmacol Exp Ther. 2011; 336(1): 9-16.doi: http://dx.doi.org/10.1124/jpet.110.170928
41. Tsai GE, Ragan P, Chang R, Chen S, Linnoila VM, Coyle JT.Increased glutamatergic neurotransmission and oxidative stress after alcohol withdrawal.Am J Psychiatry. 1998;155(6): 726-732
, 2015-03-01; accepted, 2015-03-23)

Dr. Lingjun Zuo has been working in the Department of Psychiatry, Yale University School of Medicine since 2001. She is currently the Director of the Psychiatric Genetics Lab (ZUO) in this department and a faculty member at Yale University. Her research interests are the genetics and epigenetics of psychiatric disorders and related behaviors.
酒精依赖的基于基因和基于通路的全基因组关联研究
Zuo L J , Zhang CK, Sayward FG, Cheung KH, Wang KS, Krystal JH, Zhao HY, Luo XG
基于基因的全基因组关联分析;基于通路的全基因组关联分析;细胞-细胞外基质相互作用的通路;PXN;桩蛋白;酒精依赖
Background: The organization of risk genes within signaling pathways may provide clues about the converging neurobiological effects of risk genes for alcohol dependence.Aims: Identify risk genes and risk gene pathways for alcohol dependence.Methods: We conducted a pathway-based genome-wide association study (GWAS) of alcohol dependence using a gene-set-rich analytic approach. Approximately one million genetic markers were tested in the discovery sample which included 1409 European-American (EA) alcohol dependent individuals and 1518 EA healthy comparison subjects. An additional 681 African-American (AA) cases and 508 AA healthy subjects served as the replication sample.Results: We identified several genome-wide replicable risk genes and risk pathways that were significantly associated with alcohol dependence. After applying the Bonferroni correction for multiple testing, the ‘cellextracellular matrix interactions’ pathway (p<2.0E-4 in EAs) and thePXNgene (which encodes paxillin)(p=3.9E-7 in EAs) within this pathway were the most promising risk factors for alcohol dependence. There were also two nominally replicable pathways enriched in alcohol dependence-related genes in both EAs(0.015≤p≤0.035) and AAs (0.025≤p≤0.050): the ‘Na+/Cl- dependent neurotransmitter transporters’ pathway and the ‘other glycan degradation’ pathway.Conclusions: These findings provide new evidence highlighting several genes and biological signaling processes that may be related to the risk for alcohol dependence.
[Shanghai Arch Psychiatry. 2015; 27(2): 111-118.
http://dx.doi.org/10.11919/j.issn.1002-0829.215031]
1Department of Psychiatry, Yale University School of Medicine, New Haven, CT, United States
2Department of Epidemiology and Public Health, Yale University School of Medicine, New Haven, CT, United States
3Biostatistics Resource, Keck Laboratory, Department of Genetics, Yale University School of Medicine, New Haven, CT, United States
4Center for Medical Informatics, Yale University School of Medicine, New Haven, CT, United States
5Cooperative Studies Program Coordinating Center, VA Connecticut Healthcare System, West Haven, CT, United States.
6Department of Biostatistics and Epidemiology, College of Public Health, East Tennessee State University, Johnson City, TN, United States
*correspondence: XG Luo, xingguang.luo@yale.edu; CK Zhang, kangyuzhang@hotmail.com
背景:信号通路中风险基因的构成可能可以解释酒精依赖风险基因协同的神经生物学作用。目的:识别酒精依赖的风险基因和风险基因通路。方法:我们采用基因富集(gene-set-rich)分析方法对酒精依赖进行了基于通路的全基因组关联分析(GWAS)。在包括1409名欧裔美国人(European-American,EA)酒精依赖者和1518 名EA健康对照者的探索性样本人群中检测了近一百万个基因标志物。此外,将681名非裔美国人(African-American, AA)病例和508名 AA健康受试者作为重测样本。结果:我们发现了几个与酒精依赖显著相关的可重复的全基因组风险基因和风险通路。在多重比较Bonferroni校正后,“细胞 - 细胞外基质相互作用”通路(EA样本中p<2.0E-4)和该通路中PXN基因(编码桩蛋白paxillin)(EA 样本中p=3.9E-7)是最有可能的酒精依赖的危险因素。在EA样本(0.015≤p≤0.035)和AA样本(0.025≤p≤0.050)中还有两条富含酒精依赖相关基因的可重复的通路:“Na+/ Cl-依赖性神经递质转运体”通路和“其他聚糖降解”通路。结论:一些基因和生物信号传导过程可能与酒精依赖的风险相关,本研究的发现为此提供了新的证据。
本文全文中文版从2015年06月06日起在http://dx.doi.org/10.11919/j.issn.1002-0829.215031可供免费阅览下载
猜你喜欢
杂志排行
上海精神医学的其它文章
- Unravelling psychosis: psychosocial epidemiology,mechanism, and meaning
- The transdiagnostic dimension of psychosis: implications for psychiatric nosology and research
- Finding a solution to psychosis: the emergence of a new path
- Prenatal choline and the development of schizophrenia
- Efficacy and safety of the Chinese herbal medicine shuganjieyu with and without adjunctive repetitive transcranial magnetic stimulation (rTMS) for geriatric depression: a randomized controlled trial
- Are the revised diagnostic criteria for Alzheimer’s disease useful in low- and middle-income countries?
