Identification of Cytochrome P450 (CYP) Genes in Zhikong Scallop (Chlamys farreri)
2013-04-17GUOHuihuiBAOZhenminDUHuixiaZHANGLinglingWANGShiSUNLuyangMOUXiaoyuandHUXiaoli
GUO Huihui,BAO Zhenmin,DU Huixia,ZHANG Lingling,WANG Shi,SUN Luyang,MOU Xiaoyu,and HU Xiaoli
1 Introduction
Cytochrome P450 (CYP),a group of hemeproteins,is the membership largest and function most diverse protein superfamily found in nature (Estabrook,2003).To date,more than 12000 members ofCYPbelonging to over 1000 families have been identified in living beings ranging from virus to human beings (Nelson,2011),especially in those with genome sequences.The number ofCYPgenes identified is highly variable among species.For example,57CYPgenes were found inHomo sapiens,54 inTakifugu rubripes,94 inDanio rerio,120 inStronglyocentrotus purpuratus,85 inDrosophila melanogasterand 75 inDaphnia pulex(Nelson,2009).All the members ofCYPidentified in animals cluster into 10 clans (clusters of phylogeneticly relatedCYPfamilies).For example,in vertebrate,Clan 2 includesCYPfamily 1,2,17,18 and 21; Clan 3 includesCYPfamily 3,5,6 and 9; Clan 4 includesCYPfamily 4; and the mitochondrialCYPclan includes family 11,24 and 27 (Nelson,2011).CYPsuperfamily is believed to be extremely ancient and originnate from a common ancestral gene,and consecutive duplications and subsequent divergence of the gene diversified this multigene family (Nebert and Gonzalez,1987;Nelsonet al.,1993).CYPmembers are found in all living beings ranging from prokaryotes to eukaryotes (Nebert and Gonzalez,1987).
Bivalve is one of the oldest and evolutionary most successful classes of invertebrates.From expressed sequence tags,CYPgenes have been identified in two bivalve species,mussels and oysters,which included 58CYPgenes inMytilus californianus,12 inMytilus galloprovincialis,39 inCrassostrea gigasand 14 inC.virginica(Zanetteet al.,2010).Phylogenetic analysis indicated thatCYPgenes identified inM.californianusmainly falled into Clan 2,Clan 3,Clan 4 and MitochondrialCYPclan.Clan 2 was the membership largest in deuterostomes.It is clear that identification ofCYPgenes in different bivalve species will aid to revealing the diversity ofCYPand deciphoringCYPfunction in this old invertebrate class.
As one of the most conspicuous groups of bivalve,scallops are widely distributed on the world with about over 360 species (Shumway and Parsons,2006).At present,little is known about scallopCYP(Miaoet al.,2011).Recently,transcriptome sequencing using 454 GS FLX platform and ~50-fold genome sequencing using Solexa platform have been carried out for Zhikong scallop(Chlamys farreri) (unpublished),which provided comprehensive transcriptomic and genomic resources of identifyingCYPgenes.A total of 88CYPgenes belonging to 13 families were identified in availableC.farrerisequence data with their phylogenetic relationships to human and other mollusksCYPexamined in this study.
2 Methods
2.1 Gene Identification
Significant homology toCYPwas identified by Hidden Markov Model searches (HMMER v2.3.2) (Eddy,1998)using the global PFAM model forCYP(PF00067) (http://pfam.sanger.ac.uk/) fromC.farreritranscriptome data(accession number SRA030509).The detected sequences were compared with the genomic sequence ofC.farreri(~50 fold coverage; unpublished) using BlastN with an E-value of 1e-5 as significant matching and then assembled to consensus genes.Gene identities were examined by reciprocal BLAST of the predicted genes against the NCBI non-redundant (Nr) protein database and Swiss-Prot database (http://www.ncbi.nlm.nih.gov/).
The identified sequences were translated and named based on the homology ofC.farreriCYPto those of other species using the standardized nomenclature previously assigned by Nelsonet al.(2004).Values of 40% and 55%amino acid identity were cut-offs forC.farreriCYPto be assigned to families and subfamilies,respectively.C.farreriCYPof different families were divided into clans as those in vertebrates (Nelson,2011).
2.2 Sequence Alignments and Phylogenic Analysis
All sequence alignments were performed using Muscle v3.6b (Edgar,2004),and adjusted by hand based on the MUSCLE (multiple sequence comparison by log expectation) alignment scoring function.Maximum likelihood phylogenetic trees were constructed by analyzingCYPamino acid sequences ofC.farreri,human (Nelson,2003)and other mollusks (Teunissenet al.,1992; Brownet al.,1998; Toledo-Silvaet al.,2008; Whalenet al.,2010; Miaoet al.,2011; Panet al.,2011) using MEGA (v 5.05)(Tamuraet al.,2011).The Whelan and Goldman model(WAG) of amino acid substitution with a gamma distribution of substitution rates was used (Whelan and Goldman,2001).
3 Results and Discussion
3.1 Annotation of C.farreriCYPs
Two hundred and twentyCYP-containing fragments including 85 contigs and 135 singletons were identified amongC.farreriEST.Finally,88 consensusCYPgenes were assembled manually based on BlastN results from genome sequence ofC.farreri.Forty-one of them could be translated into full-length or nearly full-lengthCYPproteins,and the others were partial.Based on the previously described rules for namingCYP(40% identity for family designation,55% identity for subfamily designation) (Nelsonet al.,2004),in this study allC.farreriCYPgenes could only be provisionally classified at level of family,soC.farreri CYPgenes assigned to oneCYPfamily were further numbered consecutively,with a ‘like’between the family number and a subordinate number in the gene name.The 88CYPs fall into 13 families based on the similarity of inferred amino acid sequences with previously namedCYP,and were mainly from Clan 2,Clan 3,Clan 4 and MitochondrialCYPclan.Four of the 88CYPfall in other clans,including 1 inCYP20 (Clan 20),2 inCYP46 (Clan 46) and 1 inCYP39 (clan not assigned) (Table 1) (Nelson,2011).
3.2 Phylogenetic Analysis of C.farreriCYPs
Maximum likelihood phylogenetic tree was constructed with the 41 full-length and nearly full-lengthC.farreri CYP,57CYPfrom human,and 10CYPfrom other mollusks,revealing the relationship amongCYPfamilies in these species (Fig.1).In general,the tree showed that 4 major clans were found inC.farreri,including Clan 2,Clan 3,Clan 4 and mitochondrialCYP.A majority ofCYPin insects,mammals and fishes also fall into these clans (Nelson,2009).
3.2.1 C.farreri CYPs in Clan 2
TheCYPmembers in Clan 2 were mainly involved in metabolism of drugs and steroids in human.Some of them could be induced through pregnane X receptor (PXR) or aryl hydrocarbon receptor (AHR) (Dograet al.,1998;Waxman,1999).As in deuterostomes and other mollusks,Clan 2 was the largest inC.farreriin which 33CYPin familyCYP1,CYP2,CYP17 andCYP356 were found.
So far,4CYP1 subfamilies (CYP1A-D) have been identified in vertebrates.A number of potentialCYP1 homologs have also been identified in deuterostomes,such asS.purpuratusandCiona intestinalis,butCYP1 have not been reported so far in protostomes (Goldstoneet al.,2007).In this study,3CYP1 genes were found inC.farreri,and 2 of them were full-length genes (member 1 and 3).The amino acid sequences of the two genes were used for phylogenetic analysis.The 2CYP1 members fromC.farrericlustered together and then clustered with those of humanCYP1 (Fig.1).In human,members ofCYP1C were not found and genes ofCYP1D members were pseudogene,and onlyCYP1A andCYP1B could be used for the construction of the phylogenetic tree(Goldstoneet al.,2009; Kawaiet al.,2010).
CYP2 is the largestCYPfamily in mammals.The members in this family were responsible for a tremendous variety of substrate oxidation (Danielson,2002).Twentythree genes inCYP2 were identified inC.farreri,accounting for 26% of the total,about 30%–49% in vertebrates and 60% in sea urchin (Goldstoneet al.,2006).All the 10 full-lengthCYP2 members ofC.farreriwere used for phylogenetic analysis,9 of them were clustered into one clade with humanCYP2 except protein 17 (Fig.1).This number 17 protein might be a member of a newCYPfamily closely relating withCYP2,which would be further classified byCYPNomenclature Committee (Nelson,2009).

Fig.1 Maximum likelihood phylogenetic tree of 41 full-length and nearly full-length members of C.farreriCYP,57 members of human CYP and 10 members of other mollusks CYP.Support values above branches are derived from 100 bootstrap replicates. cf,C.farreri; cg,C.gigas; mm,Mercenaria mercenaria; cyg,Cyphoma gibbosum; rp,Ruditapes philippinarum; ls, Lymnaea stagnalis; hs,H.sapiens.
Members inCYP17 played key roles in gonadal and adrenal steroids biosynthesis in vertebrates (Gilepet al.,2011).Only one member inCYP17 has been found in mammals,and 2 in fishes (Zhouet al.,2007).In invertebrates,4 members inCYP17 were found inHydra vulgaris(Nelson,2009) and 6 inS.purpuratus(Goldstoneet al.,2006).Three members inCYP17 were found inC.farreri,of them only member 2 was nearly full-length and could be used for phylogenetic analysis.It clustered with those in humanCYP17 andCYP21 (Fig.1).Members inCYP21 were also involved in steroid metallization,which were known to be the closest relatives to those inCYP17(Lewiset al.,1998).Members inCYP17 andCYP21 were thought to be monophyletic in molecular evolutionary researches (Lewiset al.,1998; Nelson,1999).Newly added scallop members intoCYP17 will aid to analyzing their relationships with the vertebrate homologs.
CYP356 was a family newly identified fromC.gigas.It is closely related toCYP1 andCYP17 of vertebrates(Toledo-Silvaet al.,2008).Herein,4 members ofCYP-356 were identified inC.farreri,which all clustered withCYP356A1 ofC.gigas(Fig.1).Since no members of this family had been identified in other species,CYP356 might be a bivalve specificCYPfamily that participates in special biological processes of species in this class.
3.2.2 C.farreri CYPs in Clan 3
Clan 3 consists of various members ofCYPassociated with detoxification of xenobiotics and endobiotics(Danielson,2002).Nineteen members ofCYPfromC.farreriwere assigned toCYP3,CYP5 andCYP30 of Clan 3 (Table 1).
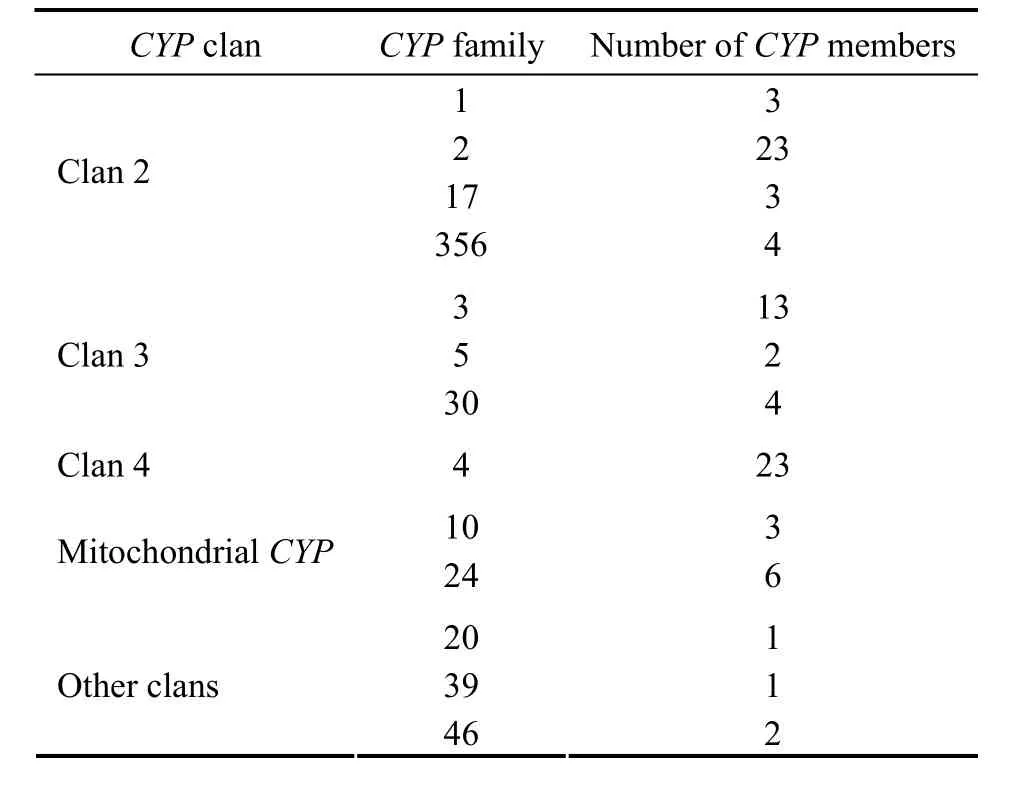
Table 1 Distribution of C.farreriCYPs in CYP clans
As one of the most importantCYPfamilies in human,members inCYP3 catalyze the metabolism of more than 50% of all clinically used drugs.Members of this family also metabolized a diverse range of other substrates such as bile acids,endogenous hormones,fungal and plant products and environmental pollutants (Danielson,2002).Genes ofCYP3 members were believed to arise from a common ancestor gene and diverged before deuterostomeprotostome splicing (Nelson,1998; Williamset al.,2004).An early deuterostomeCYP3 closely resembling the vertebrateCYP3 was reported in tunicate species,but no members ofCYP3 has been described in protostomes(Verslyckeet al.,2006).Thirteen members ofCYP3 were found inC.farreri,and all of them were partial,being not long enough for merging into phylogenetic tree.More genomic sequences are needed for phylogenetic analysis of members ofCYP3 in deuterostomes and protostomes.
Two members ofCYP5 were identified inC.farreri,and both clustered with humanCYP5A1 (Fig.1).Members ofCYP5 function as thromboxane synthase in vertebrates,and may derive from an ancestral member ofCYP3 (Verslyckeet al.,2006).So far,no members ofCYP5 have been reported in invertebrates.The identification of members ofCYP5 inC.farrerimight provide important information for revealing the evolutionary relationship ofCYP5 andCYP3.
Four members ofCYP30 were identified inC.farreri.CYP30 was a new cytochrome P450 family originally identified in gonadal tissue of the clamMercenaria mercenaria,which showed high sequence identity to mammalianCYP3 and insectCYP6,and was considered to be involved in the metabolism of steroid hormones (Brownet al.,1998).No member of this family has been reported in other species,suggesting thatCYP30 might be mollusck specific.
3.2.3 C.farreri CYPs in Clan 4
Twenty-three members ofCYPfall into Clan 4 inC.farreri,and as in vertebrates,all of them were members ofCYP4,one of the most ancientCYPfamilies in animal(Lewiset al.,1998; Nelsonet al.,2004).In vertebrates,members ofCYP4 catalyze the metabolism of fatty acids,eicosanoids and vitamin D and played important roles in chemical defense (Kirischian and Wilson,2011).Many members ofCYP4 have been identified in mollusks,some of them may be involved in fatty acid metabolism(Whalenet al.,2010).The onlyCYPmember reported in scallop was a member ofCYP4 ofC.farreri.Its expression was significantly decreased in gill and digestive gland of scallops exposing to Benzo(α)pyrene (BaP).Member 22 ofCYP4 identified in this study shared 86%amino acid similarity with it (data not shown) (Miaoet al.,2011).As shown in Fig.1,12 members documented earlier,1 ofC.farreriand 6 of other mollusks inCYP4 merged into member of humanCYP4.CYP4V10 ofCyphoma gibbosumclustered with members of humanCYP4V,the only subfamily shared by vertebrates and invertebrates (Nelson,2009).The other 18 members of molluskCYP4 analyzed in this study clustered together in a distinct clade,in which 10 members ofCYP4 identified in this study and one member ofC.farreriCYP4 reported before (Miaoet al.,2011) clustered into a subclade without members of other molluskCYP4,suggesting that this subclade may be scallop specific.Two members ofC.farreriCYP4 (7 and 14) clustered with the homologues ofC.gibbosumandRuditapes philippinarumCYP4 in another subclade,indicating these members of molluskCYP4 may function similarly.
3.2.4 Mitochondrial CYPs in C.farreri
MitochondrialCYPwas named by their subcellular locations (Nebertet al.,1991).The mitochondrialCYPs contain 3CYPfamilies in vertebrates,CYP11,CYP24 andCYP27.The members of these families involve in essential physiological functions such as metabolism of sterols,steroids and secosteroids (Danielson,2002).At least 10 families of mitochondrialCYPhave been identified in arthropods,and some members of them can metabolize xenobiotics,which is a clear difference from those of vertebrates (Danielson,2002; Feyereisen,2006; Feyereisen,2011).CYP10 from the dorsal bodies ofLymnaea stagnaliswas the only mitochondrialCYPreported in mollusks (Teunissenet al.,1992).In this study,3 members ofCYP10 and 6 ofCYP24 were found inC.farreri,and 1 member ofCYP10 and 2 ofCYP24 were assigned to phylogenetic tree.No homologue of arthropod mitochondrialCYPwas identified inC.farreri.The members of molluscanCYP10 clustered with members of humanCYP11 andCYP27,while the 2 memers ofC.farreri CYP24 clustered with humanCYP24A1 of another clade,implying that molluscanCYP10 is evolutionarily more close to CYP11 and CYP27 than CYP24 in human.
3.3 Expansion and Reduction of CYP Genes
A comparison of gene number in eachCYPclan was made amongC.farreri,D.pluex,D.melanogaster,S.purpuratus,D.rerio,T.rubripesandH.sapiens,and difference was found between deuterostomes and protostomes (Fig.2).Clan 2CYPcontained 40% to 68%members of deuterostomes (S.purpuratus,D.rerio,T.rubripesandH.sapiens)CYP,but only 8% to 38%members of protostomes (C.farreri,D.pluexandD.melanogaster)CYP,indicating that an expansion in the number of members of this clan exists in deuterostomes.Of the protostomes investigated,C.farrerihad the greatest percentage (38%) of clan 2CYPs.

Fig.2 Distribution of gene numbers in CYP clans of C.farreri,D.pluex,D.melanogaster, S.purpuratus, D.rerio,T.rubripes and H.sapiens.
Number of Clan 3 members varied from 6% to 14% of the total in deuterostomes,while from 17% to 42% in protostomes,indicating a reduction in the number of members existed in this clan in deuterostomes.There was also a relative reduction in the number of clan 4 members in deuterostomes in comparison with protostomes.The members ofCYP4 account for 8%-21% of the total in deuterostomes,while those account for 26%-49% of the total in protostomes.Further screening in the genomes of more species will aid to our understanding of the species distribution and evolution of CYP.
Acknowledgements
This study was supported by National Natural Science Foundation of China (30972239),National High-Tech R&D Program (863 Program,2012AA092204,2012AA-10A401 and 2012AA10A402),Doctoral Fund of Ministry of Education of China (20100132110014),Earmarked Fund for Modern Agro-industry Technology Research System,Natural Science Foundation of Shandong Province (ZR2009DM019),and Seed Improvement Project of Shandong Province.
Brown,D.J.,Clark,G.C.,and Beneden,R.J.V.,1998.A new cytochrome P450 (CYP30) family identi fi ed in the clam,Mercenaria mercenaria.Comparative Biochemistry and Physiology Part C,121: 351-360.
Danielson,P.B.,2002.The cytochrome P450 superfamily:Biochemistry,evolution and drug metabolism in humans.Current Drug Metabolism,3: 561-597.
Dogra,S.C.,Whitelaw,M.L.,and May,B.K.,1998.Transcriptional activation of cytochrome P450 genes by different classes of chemical inducers.Clinical and Experimental Pharmacology and Physiology,25: 1-9.
Eddy,S.,1998.Profile hidden Markov models.Bioinformatics,14: 755-763.
Edgar,R.,2004.MUSCLE: a multiple sequence alignment method with reduced time and space complexity.BMC Bioinformatics,5: 113.
Estabrook,R.W.,2003.A passion for P450s (rememberances of the early history of research on cytochrome P450).Drug Metabolism and Dispositon,31 (12): 1461-1473.
Feyereisen,R.,2006.Evolution of insect P450.Biochemical Society Transactions,34: 1252-1255.
Feyereisen,R.,2011.ArthropodCYPomes illustrate the tempo and mode in P450 evolution.Biochimica et Biophysica Acta,1814: 19-28.
Gilep,A.A.,Sushko,T.A.,and Usanov,S.A.,2011.At the crossroads of steroid hormone biosynthesis: The role,substrate specifi city and evolutionary development ofCYP17.Biochimica et Biophysica Acta,1814: 200-209.
Goldstone,J.V.,Goldstone,H.M.,Morrison,A.M.,Tarrant,A.,Kern,S.E.,Woodin,B.R.,and Stegeman,J.J.,2007.Cytochrome P4501 genes in early deuterostomes (Tunicates and Sea Urchins) and vertebrates (Chicken and Frog): Origin and diversification of theCYP1 gene family.Molecular Biology and Evolution,24 (12): 2619-2631.
Goldstone,J.V.,Hamdoun,A.,Cole,B.J.,Howard-Ashby,M.,Nebert,D.W.,Scally,M.,Dean,M.,Epel,D.,Hahn,M.E.,and Stegeman,J.J.,2006.The chemical defensome: Environmental sensing and response genes in theStrongy-locentrotus purpuratusgenome.Developmental Biology,300(1): 366-384.
Goldstone,J.V.,Jönsson,M.E.,Behrendt,L.,Woodin,B.R.,Jenny,M.J.,Nelson,D.R.,and Stegeman,J.J.,2009.Cytochrome P4501D1: A novelCYP1A-related gene that is not transcriptionally activated by PCB126 or TCDD.Archives of Biochemistry and Biophysics,482: 7-16.
Kawai,Y.K.,Ikenaka,Y.,Fujita,S.,and Ishizuka,M.,2010.TheCYP1D subfamily of genes in mammals and other vertebrates.Mammalian Genome,21: 320-329.
Kirischian,N.L.and Wilson,J.Y.,2011.Phylogenetic and functional analyses of the cytochrome P450 family 4.Molecular Phylogenetics and Evolution,62: 458-471.
Lewis,D.F.V.,Watson,E.,and Lake,B.G.,1998.Evolution of the cytochrome P450 superfamily: sequence alignments and pharmacogenetics.Mutation Research,410: 245-270.
Miao,J.,Pan,L.,Liu,N.,Xu,C.,and Zhang,L.,2011.Molecular cloning ofCYP4 and GSTpi homologues in the scallopChlamys farreriand its expression in response to Benzo[α]pyrene exposure.Marine Genomics,4: 99-108.
Nebert,D.W.,and Gonzalez,F.J.,1987.P450 genes: structure,evolution,and regulation.Annual Review of Biochemistry,56:945-993.
Nebert,D.W.,Nelson,D.R.,Coon,M.J.,Estabrook,R.W.,Feiereisen,R.,Fujii-Kuriyama,Y.,Gonzalez,F.J.,Guengerich,F.P.,Gunsalus,I.C.,Johnson,E.F.,Loper,J.C.,Sato,R.,Waterman,M.R.,and Waxman,D.J.,1991.The P450 superfamily: update on new sequences,gene mapping,and recommended nomenclature.DNA and Cell Biology,10(1): 1-14.
Nelson,D.R.,1998.Metazoan cytochrome P450 evolution.Comparative Biochemistry and Physiology Part C: Pharmacology,Toxicology and Endocrinology,121: 15-22.
Nelson,D.R.,1999.Cytochrome P450 and the Individuality of Species.Archives of Biochemistry and Biophysics,369 (1):1-10.
Nelson,D.R.,2003.Comparison of P450s from human and fugu: 420 million years of vertebrate P450 evolution.Archives of Biochemistry and Biophysics,409: 18-24.
Nelson,D.R.,2009.The cytochrome P450 homepage.Human Genomics,4: 59-65.
Nelson,D.R.,2011.Progress in tracing the evolutionary paths of cytochrome P450.Biochimica et Biophysica Acta,1814:14-18.
Nelson,D.R.,Kamataki,T.,Waxman,D.,Guengerich,F.,Estabrook,R.,Feyereisen,R.,Gonzalez,F.,Coon,M.,Gunsalus,I.,and Gotoh,O.,1993.The P450 superfamily:update on new sequences,gene mapping,accession numbers,early trivial names of enzymes,and nomenclature.DNA and Cell Biology,12 (1): 1-51.
Nelson,D.R.,Zeldin,D.C.,Hoffman,S.M.,Maltais,L.J.,Wain,H.M.,and Nebert,D.W.,2004.Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes,including nomenclature recommendations for genes,pseudogenes and alternative-splice variants.Pharmacogenetics,14: 1-18.
Pan,L.,Liu,N.,Xu,C.,and Miao,J.,2011.Identification of a novel P450 gene belonging to theCYP4 family in the clamRuditapes philippinarum,and analysis of basal- and benzo(a)pyrene-induced mRNA expression levels in selected tissues.Environmental toxicology and chemistry,32: 390-398.
Shumway,S.E.,and Parsons,G.J.,2006.Scallops: Biology,Ecology and Aquaculture.2nd edition,Elsevier,Amsterdam,651-744.
Tamura,K.,Peterson,D.,Peterson,N.,Stecher,G.,Nei,M.,and Kumar,S.,2011.MEGA5: Molecular evolutionary genetics analysis using maximum likelihood,evolutionary distance,and maximum parsimony methods.Molecular Biology and Evolution,28: 2731-2739.
Teunissen,Y.,Geraerts,W.P.M.,Heerikhuizen,H.V.,Planta,R.J.,and Joosse,J.,1992.Molecular cloning of a cDNA encoding a member of a novel cytochrome P450 family in the molluscLymnaea stagnalis.Journal of Biochemistry,112:249-252.
Toledo-Silva,G.D.,Siebert,M.,Medeiros,I.,Sincero,T.,and Moraes,M.,2008.Cloning a new cytochrome P450 isoform(CYP356A1) from oysterCrassostrea gigas.Marine Environmental Research,66: 15-18.
Verslycke,T.,Goldstone,J.V.,and Stegeman,J.J.,2006.Isolation and phylogeny of novel cytochrome P450 genes from tunicates (Cionaspp.): ACYP3 line in early deuterostomes?Molecular Phylogenetics and Evolution,40: 760-771.
Waxman,D.J.,1999.P450 gene Induction by structurally diverse xenochemicals: Central role of nuclear receptors CAR,PXR,and PPAR.Archives of Biochemistry and Biophysics,369 (1): 11-23.
Whalen,K.E.,Starczak,V.R.,Nelson,D.R.,Goldstone,J.V.,and Hahn,M.E.,2010.Cytochrome P450 diversity and induction by gorgonian allelochemicals in the marine gastropodCyphoma gibbosum.BMC Ecology,10: 24.
Whelan,S.,and Goldman,N.,2001.A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach.Molecular Biology and Evolution,18: 691-699.
Williams,E.T.,Rodin,A.S.,and Strobel,H.W.,2004.Defining relationships between the known members of the cytochrome P4503A subfamily,including five putative chimpanzee members.Molecular Phylogenetics and Evolution,33 (2):300-308.
Zanette,J.,Goldstone,J.V.,Bainy,A.C.D.,and Stegeman,J.J.,2010.Identi fi cation ofCYPgenes inMytilus(mussel) andCrassostrea(oyster) species: First approach to the full complement of cytochrome P450 genes in bivalves.Marine Environmental Research,69: S1-S3.
Zhou,L.,Wang,D.,Kobayashi,T.,Yano,A.,Paul-Prasanth,B.,Suzuki,A.,Sakai,F.,and Nagahama,Y.,2007.A novel type of P450c17 lacking the lyase activity is responsible for C21-steroid biosynthesis in the fish ovary and head kidney.Endocrinology,148 (9): 4282-4191.
杂志排行
Journal of Ocean University of China的其它文章
- Optimization of the Purification Methods for Recovery of Recombinant Growth Hormone from Paralichthys olivaceus
- Growth,Metabolism and Physiological Response of the Sea Cucumber,Apostichopus japonicus Selenka During Periods of Inactivity
- Purification and Characterization of a New Thermostable κ-Carrageenase from the Marine Bacterium Pseudoalteromonas sp. QY203
- Seasonal Changes in Food Uptake by the Sea Cucumber Apostichopus japonicus in a Farm Pond: Evidence from C and N Stable Isotopes
- What Depth Should Deep-Sea Water be Pumped up from in the South China Sea for Medicinal Research?
- Chemical Characteristics and Anticoagulant Activities of Two Sulfated Polysaccharides from Enteromorpha linza(Chlorophyta)
