An Efficient Method of Noroviruses Recovery from Oysters and Clams
2013-04-17ZHOUDeqingMALipingZHAOFengYAOLinSULaijinandLIXinguang
ZHOU Deqing,MA Liping,ZHAO FengYAO LinSU Laijin,and LI Xinguang
1) Key Laboratory for Sustainable Utilization of Marine Fisheries Resources,Ministry of Agriculture,Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences,Qingdao 266071,P.R.China
2) College of Food Science and Technology,Shanghai Ocean University,Shanghai 201306,P.R.China
1 Introduction
Noroviruses (NoVs) of theCaliciviridaefamily (Greenet al.,2000) are the major non-bacterial pathogens associated with food- and water-borne gastroenteritis in humans.Contaminated shellfish have been implicated in many outbreaks of acute gastroenteritis (Dowellet al.,1995; Kohnet al.,1995) due to their actively concentration of viruses from contaminated water (Lees,2000).Currently,NoVs remain unculturable and their detection relies exclusively on molecular biological methods.Real-time reverse transcription-polymerase chain reaction(RT-PCR) is considered to be sensitive for detection of NoVs in shellfish.However,NoVs concentrated by shellfish are difficult to identify,largely due to the insufficiency of viral recovery and/or presence of PCR inhibitors.Therefore,the most crucial step in detection of NoVs in shellfish is the viral recovery (Schultzet al.,2007),i.e.,the release and concentration of the viruses from oysters prior to (real-time) RT-PCR.Numerous protocols have been developed to solve this problem,which included glycine adsorption with or without polyethylene glycol(PEG) precipitation (Beuretet al.,2003; Hewittet al.,2006; Le Guyaderet al.,2009),proteinase K digestion(Jothikumaret al.,2005),and ultracentrifugation (Muniain-Mujikaet al.,2000).However,an optimal method has not yet been established for NoVs recovery from shellfish till present.It is necessary to identify simple and rapid methods for NoVs recovery from shellfish in order to identify the source of infection,and further contribute to the understanding of virus contaminations in shellfish and outbreak dynamics.
Bivalve molluscan shellfish such as oysters and clams filter a large volume of water in feeding,accumulating and concentrating different types of pathogens from human fecal pollutant.As a result,bivalve molluscan shellfish usually act as potential vehicles for pathogenic agents,imposing a significant health risk to human beings.According to China Fishery Statistical Yearbook (2011),oysters and clams constituted over 60% of Chinese shellfish production.This study evaluated 4 methods for recovery of NoVs in order to identify the most effective one for detection of NoVs in oysters and clams.The influences of inoculums levels and genotypes of NoVs on the recovery rate were determined by applying 2 levels of GI.3/GII.4 NoVs to oysters and clams grant tissues.Finally,the optimal method was selected out and applied to NoVs detection in oysters and clams collected from 10 coastal cities in China.
2 Materials and Methods
2.1 Virus Stocks
Stools containing GI.3 and GII.4 NoVs were provided by the Chinese Center for Disease Control and Prevention.The stools were tested by RT-PCR and NoV-positive amplicons were sequenced and identified by comparison with available sequences in GenBank using the BLAST program of the National Center for Biotechnology Information.The NoV-positive stools were 10-fold diluted with phosphate-buffered saline (PBS; 145 mmol L-1NaCl,7.7 mmolL-1Na2HPO4,and 2.3 mmolL-1NaH2PO4,pH 7.4)prior to artificial contamination and then stored at −80℃.
2.2 Artificial Contamination
Oysters (Crassostrea gigas) and clams (Ruditapes philippinarum) were purchased from a local market in Qingdao.Samples were randomly selected and kept at 4℃ for shipment.Digestive glands were dissected from oysters and clams,in which NoVs had not been previously found.A 3.0 g aliquot of the tissue sample was artificially contaminated with 10-folds dilutions of GI.3 and/or GII.4 NoV,and then incubated overnight at 4℃,allowing the attachment of GI.3 /GII.4 NoV to the homogenates.On each occasion,oyster or clam homogenates with an equal volume of PBS (pH7.4) were included as negative controls.
To calculate the recovery rate of 4 methods,2 different levels of GI.3 and GII.4 NoV were applied to oysters and clams,respectively.The high-level GI.3 and GII.4 NoV inocula,respectively,contained 1.20×106and 8.35×105NoV gene copies,while the low-level GI.3 and GII.4 NoV inocula,respectively,contained 1.50×103and 7.80×102NoV gene copies.The gene copy numbers of GI.3 and GII.4 NoVs in the inocula were determined by real-time RT-PCR.
2.3 Virus Recovery
Different concentrations of NoVs were seeded into 3.0 g of oyster/clam gland tissues.Four methods (Fig.1,A–D)were evaluated for their virus recovery efficiencies.The recovery experiment was repeated 4 times on different occasions.
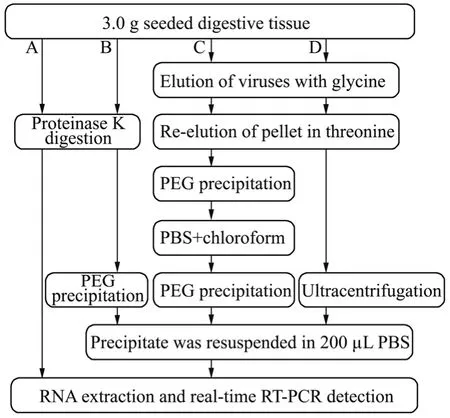
Fig.1 Principles of 4 methods (A–D) for the recovery of NoVs from shell fi sh.Method A (Jothikumar et al.,2005,with modi fi cations),Method B (newly developed in present study),Method C (Beuret et al.,2003,with modi fi cations),and Method D (Myrmel et al.,2004,with modifications).
2.3.1 Method A: Proteinase K method
Method A was slightly modified from the method originally published by Jothikumaret al.(2005).An equal amount of Tris-ethylenediamine tetraacetic acid buffer(pH 8.5) containing 100 μg mL-1proteinase K (Roche,Mannheim,Gemany) was added to 3.0 g of oyster gland tissues.The mixture was adjusted to 8.0–8.3,and then vortexed for 1 min.The mixture was incubated at 37℃for 1 h with agitation at 160 r min-1,and then heated at 65℃ for 15 min followed by centrifugation at 10000×g and 4℃ for 10 min.Thereafter,200 μL supernatant was used for RNA extraction.
2.3.2 Method B: Proteinase K-PEG 8000 method
Method B was developed on the basis of Method A.After centrifugation,the supernatant was mixed with an equal volume of 16% PEG 8000 in 0.525 mol L-1NaCl solution at 4℃ for 2 h.Then,the mixture was centrifuged at 10000 × g and 4℃ for 15 min.The pellet was resuspended in 200 μL of PBS and then used for RNA extraction.
2.3.3 Method C: Glycine-Threonine-PEG 6000 method
Method C was slightly modified from the method described by Beuretet al.(2003).Briefly,5 mL of chilled sterile 0.05 mol L-1glycine-0.14 mol L-1NaCl buffer (pH 7.5) was added to 3.0 g of tissues in 50-mL Falcon tubes.After centrifugation at 5000 × g and 4℃ for 20 min,the supernatant was collected in a second Falcon tube and stored at 4℃.The pellet was resuspended in 5 mL of 0.5 mol L-1threonine-0.14 mol L-1NaCl (pH 7.5) by vortexing for 60 s.After centrifugation at 5000 × g and 4℃ for 20 min,the supernatant was combined with the first supernatant in a third Falcon tube,and the pellet was discarded.Subsequently,10 mL of 12% PEG 6000 in 0.3 molL-1NaCl solution (4℃) was added,and the suspension was allowed to precipitate at 4℃ for 2 h.The resulting precipitate was centrifuged at 6700 × g and 4℃ for 30 min and resuspended in 5 mL of PBS (pH 7.5).The pellet was re-suspended in 5 mL of chloroform by vortexing for 60 s,and then centrifuged at 2000 × g and 4℃ for 30 min.The supernatant was reprecipitated with 5 mL of 12% PEG 6000 in 0.3 mol L-1NaCl solution (4℃) at 4℃ for 2 h.After centrifugation at 10000 × g and 4℃ for 15 min,the pellets were resuspended in 200 μL of PBS and then directly used for RNA extraction.
2.3.4 Method D: Glycine-Threonine-Ultracentrifugation method
The Method D was slightly modified from the method described by Myrmelet al.(2004).The virus elution process was the same as that in Method C.After the second centrifugation at 5000 × g for 20 min,the combined supernatant was collected and centrifuged at 190000 × g and 4℃ for 2 h (Hitachi Himac CP 100 WX Ultracentrifuge,Rotor P100 AT2).The pellet was resuspended in 200 μL of PBS and then used for RNA extraction.
2.4 Viral RNA Extraction
Viral RNA was extracted using the High Pure Viral Nucleic Acid Kit (Roche,Mannheim,Germany) following the manufacturer’s instructions.
2.5 Real-time RT-PCR Detection of NoVs
To allow an accurate estimation of the NoV gene copy number,the transcript was generatedin vitro.The PCR products flanked by primer COG2R and COG2F and primer COGR and COGF were directly cloned into pGEM-T vector (Promega,USA) according to the manufacturer’s instructions.Clones were sequenced (Invitrogen,Shanghai,China) to ensure the sequence integrity.Plasmids with the correct inserts were linearized by digestion withNdeI (Takara,China),and the linear DNA was used as the template for run-off transcripts using T7 RiboMAXTM Express Large Scale RNA Production System kit (Promega,USA).Transcript RNA was resolved in nuclease-free water and frozen at −80℃ until use.Transcript RNA concentration was determined using a NanoDrop ND-2000 (Thermo) spectrophotometer.
The GII NoVs were detected using the primer COG2R and COG2F and the probe RING II,and the GI NoVs detected using the primer COGR and COGF and the probe RING I (Kageyamaet al.,2003).
Real-time RT-PCR assay was carried out using the one-step system (TaKaRa,China) in duplicate 20 μL reaction mixtures containing 2 μL of extracted RNA,200 nmol L-1GI or GII primers,and 400 nmol L-1probes.The RT-PCR was performed with a Lightcycler 2.0 (Roche,Germany) under the following conditions: reverse transcription at 42℃ for 5 min,denaturation at 95℃ for 10 s,and 45 cycles of amplification with denaturation at 95℃for 15 s and annealing and extending at 52℃ for 30 s.RNA transcripts of GI.3 or GII.4 NoVs were used as positive controls and non-spiked samples used as negative controls.
2.6 Oyster and Clam Samples
Between January 2011 and February 2012,a total of 84 oysters and 86 clams were collected from 10 seafood markets in different cities,including Dalian,Laizhou,Yantai,Weihai,Qingdao,Rizhao,Lianyungang,Zhoushan,Xiamen and Guangzhou.Samples were packaged individually,randomly selected,and kept on ice during the shipment.The samples were immediately shucked upon the arrival to the laboratory.Digestive gland tissues were separated from whole body,cut into small portions,homogenized and divided into 3.0-g portions.Samples were analyzed following 2.3 (Method B),2.4,and 2.5.
2.7 Data Analysis
The recovery efficiency was calculated per individual method as ‘the mean recovered gene copy number of GI.3 and GII.4 NoV’ per ‘mean inoculated gene copy number of GI.3 or GII.4 NoVs’.
Statistical analysis was performed using SPSS 17.0 with the non-parametric Kruskal-Wallis test (KW-test).Significance levels were set at 0.05.
3 Results
3.1 Recovery Efficiency of 4 Methods in Enrichment of GI.3 NoV
Table 1 showed that the efficiency of Method B was the highest for recovery of GI.3 NoVs from spiked oyster digestive tissues,with 100% positive RT-PCR reactions(16/16).The mean recovery efficiency of method B was higher than that of other 3 methods with both high and low levels of inocula (Fig.2).For clams,Method D successfully recovered GI.3 NoVs in all PCR reactions,but its recovery efficiency was lower than that of Method B.Taking into account the influence of different shellfish species on 4 recovery methods,we concluded that all the 4 methods were more efficient in recovery of NoVs from clams than from oysters,but there was a lack of significant differences between these 2 shellfish species.
For recovery of GI.3 NoVs,these 4 methods showed no significant differences in effeciency (P>0.05).However,there were significant differences between methods B and C as well as methods B and D (P=0.037).
In general,Method B yielded the maximum amount of positive NoV gene copy numbers (30/32) in both treatments,followed by methods A,D and C.

Table 1 Efficiency of 4 methods for recovery of GI.3 NoVs from oysters and clams
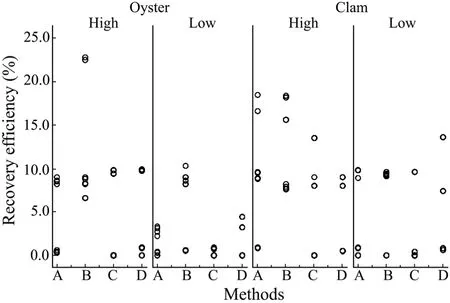
Fig.2 The variation in GI.3 NoV recovery efficiency with high (1.20×106 copies) and low (1.50×103 copies) levels of virus inocula of shellfish samples.Each circle represents a PCR reaction.
3.2 Recovery Efficiency of 4 Methods for Enriching GII.4 NoV
Similar patterns were observed in the detection of spiked levels of GII.4 NoVs in oysters and clams.The recovery efficiency was higher with a high level of GII.4 NoV inocula compared to that with a low level of inocula(Fig.3).Methods A and B were found more efficient and sensitive for estimating the recovery efficiency of GII.4 NoVs from spiked oysters digestive tissue (>1% in all PCR reactions) (Fig.3).However,Method B yielded the highest mean recovery efficiency,i.e.,16.4% for oysters and 12.8% for clams with the high level inocula,and 9.8% for oysters and 11.8% for clams with the low level inocula (Table 2).
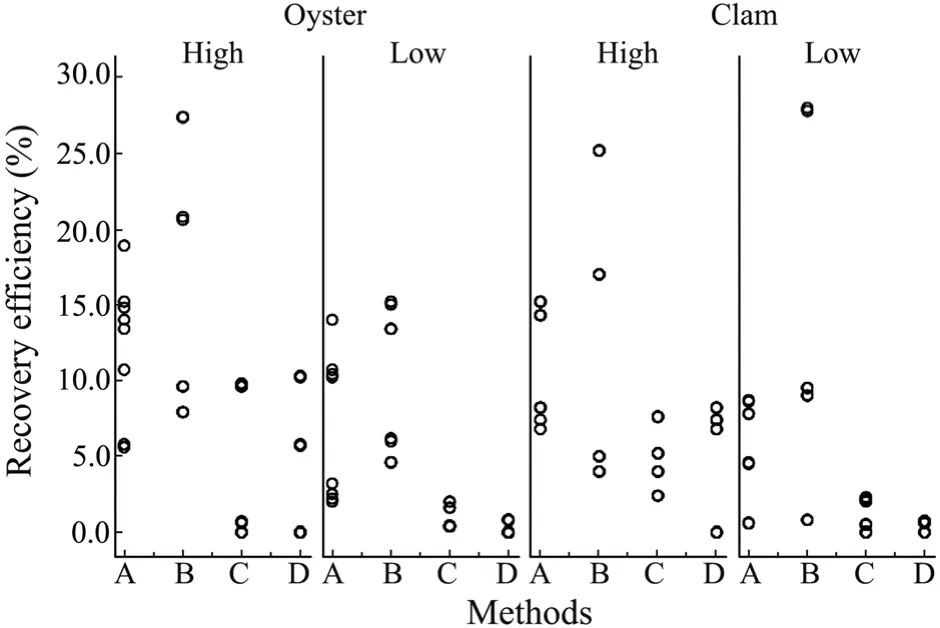
Fig.3 The variations in GII.4 NoV recovery efficiency with high (8.35×105 copies) and low (7.80×102 copies)levels of virus inocula of shellfish samples.Each circle represents a PCR reaction.
Unlike that of GI.3 NoVs,the recovery of GII.4 NoVs showed significant differences in the mean recovery efficiency among 4 methods (P=0.01).

Table 2 The efficiency of 4 methods for recovery of GII.4 NoVs from oysters and clams samples
3.3 Comparison of 4 Methods for Recovery of GI.3 and GII.4 NoVs
The genotype of NoVs had no significant influence on the respective recovery efficiency (KW-test;P>0.05).However,there was significant difference in the mean recovery efficiency among 4 methods for both oysters and clams (P<0.05).Compared with methods C and D,the Method B was more feasible and advantageous.The inoculation levels of GI.3 and GII.4 NoVs significantly affected the recovery efficiency of different methods(KW-test;P=0.037).The recovery efficiency was higher with a higher level of inocula.
Together,the results indicate that Method B was more efficient,which allowed an overall sensitive and repeatable recovery of both GI.3 and GII.4 NoVs from the oysters and clams digestive tissue.Hence,Method B was chosen for subsequent investigation on NoV pollution of oysters and clams.
3.4 Presence of NoVs in Oysters and Clams
NoVs were detected in both oysters and clams samples.The average detection rate of NoVs in the two shellfish species was 8.82% (15/170).The isolation rates of NoVs were 10.47% (9/86) in clams and 7.14% (6/84) in oysters.All detected NoVs are of genotype GII.12.
4 Discussion
The NoVs are genetically and antigenically diverse(Atmaret al.,2006).Their genetic classification system is based on the relatedness of the complete VP1 capsid pro-tein,and currently there are 5 recognized genogroups(Zhenget al.,2006).Of these,GI,GII,and GIV strains may infect humans,whereas GIII and GV strains may infect cows and mice,respectively.For some years,GII strains,particularly those of the GII.4 cluster,were the predominant viruses detected worldwide (Siebengaet al.,2009).Other strains,especially GI strains,were more often transmitted via food or environmental contamination (Lysenet al.,2009; Nodaet al.,2008).Recent research has shown that NoVs can specifically bind to antigens in the oyster gut that are similar to human blood group antigens (HBGAs) (Le Guyaderet al.,2006; Tianet al.,2006).The genetic diversity of NoVs is also reflected in their binding capacity to various HBGA structures.The differences observed between GI.1 and GII.4 binding to human HBGAs were also present in oyster tissues (Maaloufet al.,2010).In the present study,the genotype of NoVs had no significant influence on the respective recovery efficiency of 4 methods,possibly due to the way of NoVs artificially attached in shellfish digestive tissues was different from that of NoVs binding to the HBGA structures.
During sample preparation,the gills,mantle,connective tissue,and other tissues were cut off,leaving the digestive tissue as the target.Most frequently used methods have focused on the dissected bivalve digestive diverticulum (digestive gland) as the starting material for virus extraction.This organ has been shown to be the target of contamination within the bivalve (Romaldeet al.,1994).Digestive tissues comprise approximately 10% of the body mass of the bivalve but contain a large majority of the contaminating virus.Thus,targeting the digestive gland avoids the need to process tissues that contain small amounts of virus with abundant potential PCR inhibitors,thereby reducing the processing time and improving the sensitivity and quality of the extraction.
A number of publications have detailed several approaches in the treatment of bivalve digestive glands for the release,concentration,and purification of viruses,which included acid adsorption-elution (Jaykuset al.,1996),direct glycine buffer elution (Leeset al.,1994),immunomagnetic bead extraction (Parket al.,2008; Yaoet al.,2009),virus precipitation using Cat-Floc (Richardset al.,1982) or PEG (Jaykuset al.,1996),and solvent extraction using chloroform (Atmaret al.,1993; Mullendoreet al.,2001) or chloroform/butanol (Atmaret al.,1995).Most of these methods are time-consuming,which may cause the loss of viral genomes during successive steps (Grif fi net al.,2003).The ultracentrifugation-based approaches can be used to pellet NoVs (Rzeżutkaet al.,2008) but requires expensive equipment.Of the 4 methods used in the present study,proteinase K digestion was rapid with less manual steps and easy to standardize or adapt to different quantities of shell fi sh.Our results showed that methods A and B had higher recovery efficiency compared with methods C and D,even with a low level of virus inoculation.Proteinase K is efficient in releasing NoVs from shellfish.Its superiority has been reported by Comelliet al.(2008) and Uhrbrandet al.(2010).However,we found Method B had higher recovery efficiency than Method A.The precipitation of PEG 8000 may have improved the detection limits in some instances.
Until recently,data of the NoV pollution have been limited and GI NoVs have not been detected in retail shellfish in China (Liuet al.,2009).In the present study,NoV contamination of oysters and clams from seafood retail markets of 10 coastal cities in China is described.Results provide experimental data for risk assessment of virus-related shellfish-borne hazards to the shellfish consumers.We detected viral contamination in both oysters and clams (mean detection rate 8.82%).Thus,it is necessary to develop a standard method for determination of the extent of NoV contamination of shellfish in retail markets.All NoVs from positive samples were found of the genotype GII.12,indicating that GII.12 is the major genotype in shellfish in China.However,GII.4 has been reported as the predominate cause of NoV infections worldwide (Siebengaet al.,2009).Further study is needed to confirm if GII.12 had become a new prevalent epidemic strain in China as well as other countries.
In conclusion,the proteinase K-PEG 8000 method(Method B) has significantly improved the NoV detection limit compared with the proteinase K digestion method(Method A).The former was successfully applied to oysters and clams samples for NoV detection and was shown to be a good candidate method for recovering NoVs from shellfish.
Acknowledgements
This work was supported by the China-Australia Bilateral Research Program (No.2010DFA31720) and the National Key Technology R&D Program (2012BAD28B05).
Atmar,R.L.,Metcalf,T.G.,Neill,F.H.,and Estes,M.K.,1993.Detection of enteric viruses in oysters by using the polymerase chain reaction.Applied and Environmental Microbiology,59 (2): 631-635.
Atmar,R.L.,Neill,F.H.,Romalde,J.L.,Le Guyader,F.,Woodley,C.M.,Metcalf,T.G.,and Estes,M.K.,1995.Detection of Norwalk virus and hepatitis A virus in shellfish tissues with the PCR.Applied and Environmental Microbiology,61: 3014-3018.
Atmar,R.L.,and Estes,M.K.,2006.The epidemiologic and clinical importance of norovirus infection.Gastroenterology Clinics of North America,35: 275-290.
Beuret,C.,Baumgartner,A.,and Schluep,J.,2003.Virus-contaminated oysters: a three-month monitoring of oysters imported to Switzerland.Applied and Environmental Microbiology,69: 2292-2297.
Comelli,H.L.,Rimstad,E.,Larsen,S.,and Myrmel,M.,2008.Detection of norovirus genotype I.3b and II.4 in bioaccumulated bluemussels using different virus recovery methods.International Journal of Food Microbiology,127: 53-59.
Dowell,S.F.,Groves,C.,Kirkland,K.B.,Cicirello,H.G.,Ando,T.,Jin,Q.,Gentsch,J.R.,Monroe,S.S.,Humphrey,C.D.,Slemp,C.,Dwyer,D.M.,Meriwether,R.A.,and Glass,R.I.,1995.A multistate outbreak of oyster-associated gastroen-teritis: implications for interstate tracing of contaminated shell fi sh.Journal of Infectious Diseases,171: 1497-1503.
Green,K.Y.,Ando,T.,Balayan,M.S.,Berke,T.,Clarke,I.N.,Estes,M.K.,Matson,D.O.,Nakata,S.,Neill,J.D.,Studdert,M.J.,and Thiel,H.J.,2000.Taxonomy of the caliciviruses.Journal of Infectious Diseases,181: S322-330.
Grif fi n,D.W.,Donaldson,K.A.,Paul,J.H.,and Rose,J.B.,2003.Pathogenic human viruses in coastal waters.Clinical Microbiology Reviews,16 (1): 129-143.
Hewitt,J.,and Greening,G.E.,2006.Effect of heat treatment on hepatitis a virus and norovirus in New Zealand greenshell mussels (Perna canaliculus) by quantitative real-time reverse transcription PCR and cell culture.Journal of Food Protection,69: 2217-2223.
Jaykus,L.A.,DeLeon,R.,and Sobsey,M.D.,1996.A virion concentration method for detection of human enteric viruses in oysters by PCR and oligoprobe hybridization.Applied and Environmental Microbiology,62: 2074-2080.
Jothikumar,N.,Lowther,J.A.,Henshilwood,K.,Lees,D.N.,Hill,V.R.,and Vinje,J.,2005.Rapid and sensitive detection of noroviruses by using TaqMan-based one-step reverse transcription-PCR assays and application to naturally contaminated shell fi sh samples.Applied and Environmental Microbiology,71: 1870-1875.
Kageyama,T.,Kojima,S.,Shinohara,M.,Uchida,K.,Fukhusi,S.,Hoshino,F.B.,Takeda,N.,and Katayama,K.,2003.Broadly reactive and highly sensitive assay for Norwalk-like viruses based on realtime quantitative reverse transcription-PCR.Journal of Clinical Microbiology,41: 1548-1557.
Kohn,M.A.,Furley,T.A.,Ando,T.,Curtis,M.,Wilson,S.A.,Jin,Q.,Monroe,S.S.,Baron,R.C.,McFarland,L.M.,and Glass,R.I.,1995.An outbreak of Norwalk virus gastroenteritis associated with eating raw oysters.Implications for maintaining safe oyster beds.Journal of American Medical Association,273: 466-471.
Lees,D.N.,Henshilwood,K.,and Dore,W.J.,1994.Development of amethod for detection of enteroviruses in shell fi sh by PCR with poliovirus as a model.Applied and Environmental Microbiology,61: 2999-3005.
Lees,D.N.,2000.Viruses and bivalve shellfish.International Journal of Food Microbiology,59: 81-116.
Le Guyader,F.,Loisy,F.,Atmar,R.L.,Hutson,A.M.,Estes,M.K.,Ruvoën-Clouet,N.,Pommepuy,M.,and Le Pendu,J.,2006.Norwalk virus-speci fi c binding to oyster digestive tissues.Emerging Infectious Diseases,12: 931-936.
Le Guyader,F.,Parnaudeau1,S.,Schaeffer,J.,Bosch,A.,Loisy,F.,Pommepuy,M.,and Atmar,R.L.,2009.Detection and quantification of Noroviruses in shellfish.Applied and Environmental Microbiology,75: 618-624.
Liu,S.F.,Li,Z.,and Zhou,D.Q.,2009.Epidemic of noroviruses in bivalves in Qingdao.Progress in Fishery Sciences,30: 61-66.
Lysen,M.,Thorhagen,M.,Brytting,M.,Hjertqvist,M.,Andersson,Y.,and Hedlund,K.O.,2009.Genetic diversity among food-borne and waterborne norovirus strains causing outbreak in Sweden.Journal of Clinical Microbiology,47: 2411-2418.
Maalouf,H.,Zakhour,M.,Le Pendu,J.,Le Saux,J.C.,Atmar,R.L.,and Le Guyader,F.S.,2010.Distribution in tissue and seasonal variation of norovirus genogroup I and II ligands in oysters.Applied and Environmental Microbiology,76 (16):5621-5630.
Mullendore,J.L.,Sobsey,M.D.,and Shieh,Y.C.,2001.Improved method for the recovery of hepatitis A virus from oysters.Journal of Virological Methods,94: 25-35.
Muniain-Mujika,I.,Girones,R.,and Lucena,F.,2000.Viral contamination of shell fi sh: evaluation of methods and analysis of bacteriophages and human viruses.Journal of Virological Methods,89: 109-118.
Myrmel,M.,Berg,E.M.,Rimstad,E.,and Grinde,B.,2004.Detection of enteric viruses in shellfish from the Norwegian coast.Applied and Environmental Microbiology,70 (5): 2678-2684.
Noda,M.,Fukuda,S.,and Nishio,O.,2008.Statistical analysis of attack rate in norovirus foodborne outbreaks.International Journal of Food Microbiology,122: 216-220.
Park,Y.,Cho,Y.H.,Jee,Y.,and Ko,G.,2008.Immunomagnetic separation combined with real-time reverse transcriptase PCR assays for detection of norovirus in contaminated food.Applied and Environmental Microbiology,74: 4226-4230.
Richards,G.P.,Goldmintz,D.,Green,D.L.,and Babinchak,J.A.,1982.Rapid methods for extraction and concentration of poliovirus from oyster tissues.Journal of Virological Methods,5: 285-291.
Romalde,J.L.,Estes,M.K.,Szucs,G.,Atmar,R.L.,Woodley,C.M.,and Metcalf,T.G.,1994.In situdetection of hepatitis A virus in cell cultures and shell fi sh tissues.Applied and Environmental Microbiology,60: 1921-1926.
Rzeżutka,A.,Chrobocińska,M.,Kaupke,A.,and Mizak,B.,2008.Application of an ultracentrifugation-based method for detection of feline calicivirus (a Norovirus Surrogate) in experimentally contaminated delicatessen meat samples.Food Analytical Methods,1: 56-60.
Schultz,A.C.,Saadbye,P.,Hoorfar,J.,and Norrung,B.,2007.Comparison of methods for detection of norovirus in oysters.International Journal of Food Microbiology,114: 352-356.
Siebenga,J.,Vennema,H.,Zheng,D.P.,Vinje,J.,Lee,B.E.,Pang,X.L.,Ho,E.C.M.,Lim,W.,Choudekar,A.,Broor,S.,Halperin,T.,Rasool,N.B.G.,Hewitt,J.,Greening,G.E.,Jin,M.,Duan,Z.J.,Lucero,Y.,O’Ryan,M.,Hoehne,M.,Schreier,E.,Ratcliff,R.M.,White,R.A.,Iritani,N.,Reuter,G.,and Koopmans,M.,2009.Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants,2001-2007.Journal of Infectious Diseases,200: 802-812.
Tian,P.,Bates,A.H.,Jensen,H.M.,and Mandrell,R.E.,2006.Norovirus binds to blood group A-like antigens in oyster gastrointes-tinal cells.Letters in Applied Microbiology,43: 645-651.
Uhrbrand,K.,Myrmel,M.,Maunula,L.,Vainio,K.,Trebbien,R.,Nørrung,B.,and Schultz,A.C.,2010.Evaluation of a rapid method for recovery of norovirus and hepatitis A virus from oysters and blue mussels.Journal of Virological Methods,169: 70-78.
Yao,L.,Wu,Q.,Wang,D.,Kou,X.,and Zhang,J.,2009.Development of monoclonal antibody-coated immunomagnetic beads for separation and detection of norovirus (genogroup II)in faecal extract samples.Letters in Applied Microbiology,49:173-178.
Zheng,D.P.,Ando,T.,Fankhauser,R.L.,Beard,R.S.,Glass,R.,and Monroe,S.S.,2006.Norovirus classification and proposed strain nomenclature.Virology,346: 312-323.
杂志排行
Journal of Ocean University of China的其它文章
- Optimization of the Purification Methods for Recovery of Recombinant Growth Hormone from Paralichthys olivaceus
- Growth,Metabolism and Physiological Response of the Sea Cucumber,Apostichopus japonicus Selenka During Periods of Inactivity
- Purification and Characterization of a New Thermostable κ-Carrageenase from the Marine Bacterium Pseudoalteromonas sp. QY203
- Seasonal Changes in Food Uptake by the Sea Cucumber Apostichopus japonicus in a Farm Pond: Evidence from C and N Stable Isotopes
- What Depth Should Deep-Sea Water be Pumped up from in the South China Sea for Medicinal Research?
- Chemical Characteristics and Anticoagulant Activities of Two Sulfated Polysaccharides from Enteromorpha linza(Chlorophyta)
