New insight into the mechanism of DNA polymerase I revealed by single-molecule FRET studies of Klenow fragment
2022-08-31RokshanaParvinQiJia贾棋JianbingMa马建兵ChunhuaXu徐春华YingLu陆颖FangfuYe叶方富andMingLi李明
Rokshana Parvin Qi Jia(贾棋) Jianbing Ma(马建兵) Chunhua Xu(徐春华)Ying Lu(陆颖) Fangfu Ye(叶方富) and Ming Li(李明)
1Beijing National Laboratory for Condensed Matter Physics,Institute of Physics,Chinese Academy of Sciences,Beijing 100190,China
2School of Physical Sciences,University of Chinese Academy of Sciences,Beijing 100049,China
Keywords: smFRET,Klenow fragment,GC-richness,strand displacement
1. Introduction
Interactions between DNA and protein complexes are the center of essential cellular processes such as transcription,cell division, metabolism, and development of organisms.[1]DNA polymerases (DNA pols) play a vital role in DNA replication[2,3]and repair.[4]DNA pols have been utilized in many molecular biological applications like polymerase chain reactions(PCR)[5,6]and DNA sequencing.[7]Escherichia coliDNA pol I (Klenow fragment [KF]), a multifunctional active truncated proteolytic form of polymerase I,consists of a polymerase active site(5′→3′)that incorporates nucleotides and a 3′→5′exonuclease active site that excises wrongly incorporated nucleotides, and it does not contain any 5′→3′exonuclease active site. This property of the KF makes it very suitable for use in manipulating DNA fragments in a test tube.It is extremely useful in molecular cloning and DNA labeling experiments,and also in a variety of techniques to achieve insight into mechanisms of different polymerases and nucleic acid—protein interactions.
In this article, we study how the richness of GC pairs of DNA chains may influence the interaction between KF and DNA. It has been well known that GC-rich DNA sequences have importance in DNA replication, transcription, cell cycle control, methylation, chromatin structure, and development.Sequences with more GC-content are more stable than sequences with less GC-content. This stability is mainly due to base stacking interactions.[8]When GC-rich constructs form secondary structures, particularly hairpin and loops, they are very stable and do not melt well at usual PCR denaturation temperatures.[9,10]Here we are interested in how this stability may influence the strand displacement mechanism of Klenow fragment.
We use single molecule fluorescence resonance energy transfer (smFRET) and adopt the newly-developed D-loopbased technique[11]to investigate the mechanism of KF. sm-FRET is a powerful[12–14]and widely used[13,15]technique for studying biological molecules,their conformation and conformational dynamics,[16]and detailed kinetics of complex biochemical reactions, particularly, those between proteins and nucleic acids. It acts as a spectroscopic ruler,[14,17]which exploits strong donor–acceptor distance-dependence to resolve distance changes at nanometer scale.[18]The newly-developed D-loop-based technique is used to improve resolution for KFDNA interaction results throughout smFRET measurements.Our assay enables us to investigate the dynamics of strand displacement by KF in real time,and our results reveals that KF may backslide during the strand displacement and the probability of backsliding is strongly influenced by the GC-richness of DNA sequences.
2. Materials and methods
2.1. Purification and labeling of DNA substrates
To prepare the DNA substrates, all the required oligonucleotides were purchased from Sangon Biotech Ltd. (Shanghai, China). The double-stranded DNA molecules were annealed by incubating the mixture at 95◦C for 5 minutes, and then slowly cooled down in room temperature for 7 hours.The annealing was carried out in an annealing buffer containing 100-mM NaCl, 25-mM Tris-HCl, pH 7.5. A 2% agarose gel was used to purify the dsDNA to ensure the absence of free single-stranded DNA. The DNAs were labeled with Cy3 and Cy5 dyes.
2.2. Klenow fragment(3′→5′exo-)
We used KF(3′→5′exo-)which is an N-terminal truncation of DNA pol I that retains 5′→3′polymerase activity,but lacks the 5′→3′exonuclease activity and has two mutations(D355A,E357A)which abolish the 3′→5′exonuclease activity.[19,20]The elimination of exonuclease activity makes KF exo- the enzyme of choice for biotin labeling of DNA probes by random primed method,[21]and for DNA sequencing by the Sanger dideoxy method.[22]The Klenow fragments(200 units,5000 U/mL)were purchased from NEB(New England Biolabs,Inc.).
2.3. Buffers
The 10× NE Buffer2 was purchased from NEB (New England Biolabs, Inc.), and the 1× Buffer2 contains 50-mM NaCl,10-mM Tris-HCl,10-mM MgCl2,1-mM DTT,pH 7.9.The T50 buffer contains 50-mM NaCl, 20-mM Tris-HCl,pH 7.5. A dNTP mixture(2.5-mM each)was purchased from TaKaRa Bio USA,Inc.
2.4. Single molecule measurements for KF
The single molecule FRET measurements were carried out with a home-built objective type total internal reflection fluorescence(TIRF)microscope. An oil immersion objective(100×,N.A.1.49)was used to create an evanescent field of illumination. Cy3 was excited by a 532-nm sapphire laser(Coherent,Inc.USA).The fluorescence signals from Cy3 and Cy5 were separated by a dichroic mirror, and were collected by an electron multiplying charge-coupled device camera(iXON,Andor technology,South Windsor,CT,USA).The coverslips(Fisher Scientific, USA) and slides were cleaned throughout a process by using acetone, methanol, a mixture of sulfuric acid and hydrogen per oxide at a ratio of 7:3, a mixture of sodium hydroxide and ethanol, and finally with a mixture of 47.5-mL methanol, 2.5-mL acetic acid and 0.5-mL APTES(Roche Diagnostics). The coverslips were coated with a mixture of 99% mPEG-SVA (MW 5000, Laysan Bio, Inc.) and 1% biotin-PEG-SVA (MW 5000, Laysan Bio, Inc.). Streptavidin was applied to the micro chamber before the immobilization of biotinylated DNA, and was incubated for 10 minutes.After washing,20-pM–70-pM DNA was added into the chamber and incubated for 5 minutes. After removal of free DNA molecules with 200-µL T50 buffer,an oxygen scavenging system(0.8%D-glucose, 1-mg/mL glucose oxidase, 0.4-mg/mL catalase, and 1-mM Trolox) was added to the reaction buffer and then flowed into the chamber to initiate imaging. Then,10 unit of KF with reaction buffer were added into the chamber for KF-DNA binding reaction,and imaging was initiated.After the initiation of imaging,KF and dNTP were flowed into the chamber. All the single molecule measurements were carried out at an exposure time of 50 ms–100 ms and a constant temperature of 22◦C.The FRET efficiency was calculated asIA/(ID+IA), whereIDandIAare the corrected emission intensities of the donor and acceptor,respectively.
3. Results
3.1. Strand displacement on Y-shaped DNA
A home-built total internal reflection fluorescence(TIRF)microscope was used to monitor the interaction between unlabeled KF and donor-acceptor labeled DNA. We start with monitoring the interaction between KF and Y-shaped DNA.As shown in Fig.1(a),the Y-shaped DNA is formed by 3 strands of ssDNA. The FRET donor (Cy3) was labeled at the primer and the FRET acceptor (Cy5) was labeled at the opening of DNA fork. Strand displacement of KF will unwind the DNA fork and increase the distance between Cy3 and Cy5, leading to the decrease of the FRET value;on the other hand,the backsliding of KF increases the FRET value. Therefore we can monitor the dynamics of strand displacement by measuring the FRET value changes.
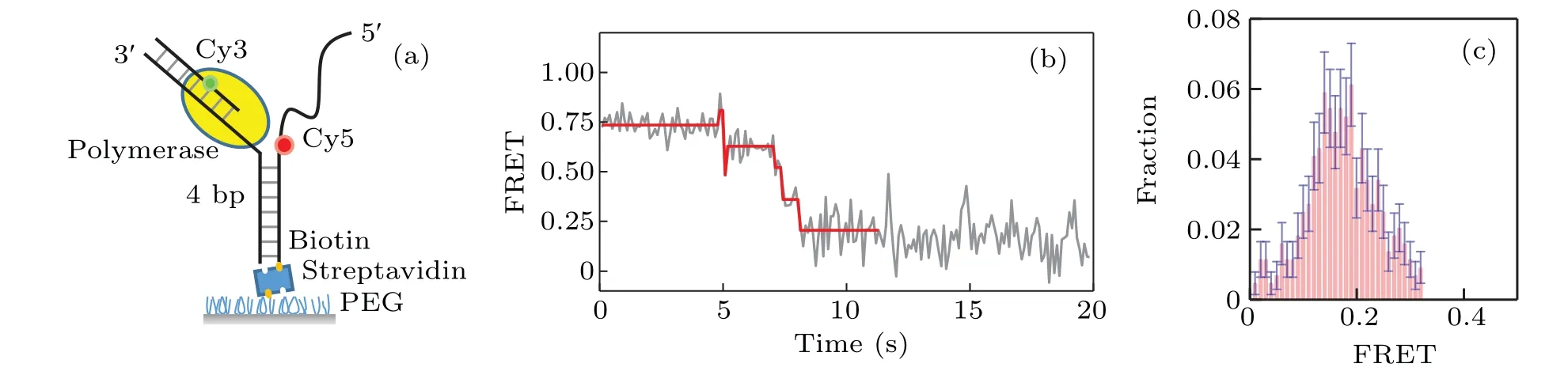
Fig.1. Strand displacement on Y-shaped DNA.(a)Cy3(green)and Cy5(red)labeled Y-shaped DNA;(b)typical trace for strand displacement;(c)distribution of the ending FRET value of strand displacement by KF on Y-shaped DNA.
Figure 1(b)gives a typical FRET trace resulting from the strand displacement. We can see there exist several jumps of the FRET value change.Most of them are from a higher FRET value to a lower one, which correspond to the unwinding behavior of the DNA by KF.As KF continuously unwinds DNA,the FRET traces decrease to a value∼0.2. The distribution of the ending values of the FRET traces is given in Fig.1(c).Note that there also exist in the FRET traces very few jumps from lower FRET values to higher ones, which probably represent the backsliding of KF on the DNA.
3.2. Strand displacement on D-loop DNA
We proceed to study the interaction between KF and Dloop DNA As shown in Fig.2(a),the arc of the D-loop DNA molecules we use is a 60-bp long dsDNA,with the string being consisted of a 31-nt DNA.A 25-mer primer sequence-(5′-TGTACATTCAATCACCGACG(T)TTCC-3′) is labeled with Cy3 FRET donor. The biotin group is labeled at 5′-PHOCy5 FRET acceptor is labeled at the 5′end of the 60-bp ds-DNA. The estimated force of the D-loop is about 7 pN. A long hairpin of 100 bp is attached to the D-loop to ensure the full activity of KF. Figure 2(b) gives a typical FRET trace of strand displacement result. After the binding of KF,the FRET value is increased from 0.82 to 0.96; and it is clearly shown that there exist both forward and backward steps,which represent the unwinding and backsliding phenomena by KF,respectively. Figure 2(c) gives the distribution of the ending FRET value. Note that the peak of the distribution locates around 0.08, which is much lower than the corresponding value in the case of Y-shaped DNA. This lowering of the FRET ending value indicates that the D-loop structure helps to enhance the resolution and efficiency of smFRET in studying KF-DNA interaction.
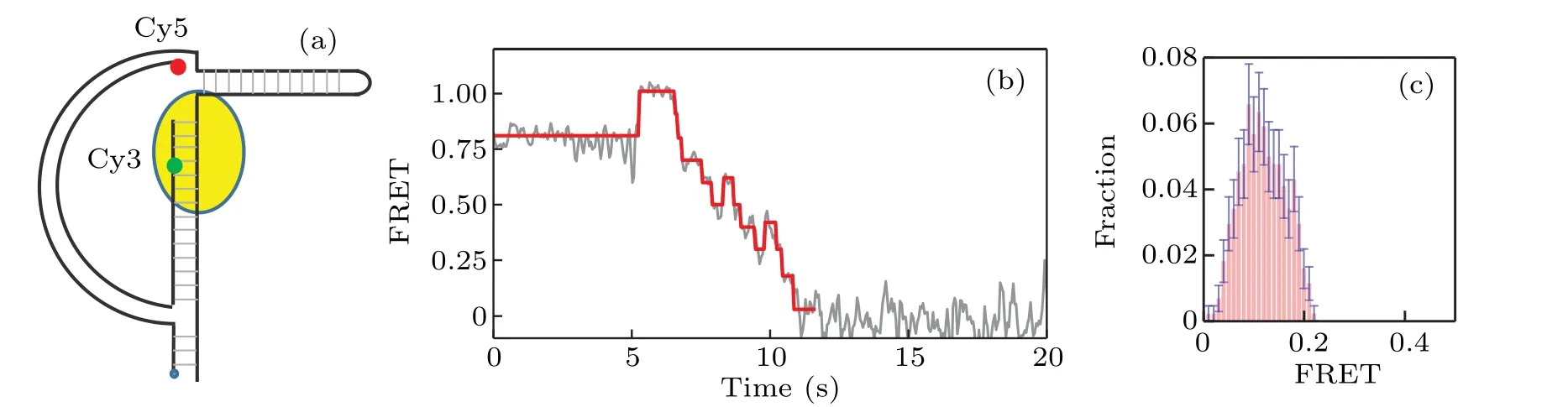
Fig. 2. Strand displacement on D-loop DNA. (a) Structure of D-loop DNA labeled with Cy3 (green) and Cy5 (red); (b) typical trace for strand displacement,where backward steps are more distinct than those in Y-shaped DNA;(c)distribution of the ending FRET value of strand displacement by KF on D-loop DNA.
3.3. Influence of GC-pair richness on the strand displacement by KF
Because GC-rich sequences are important in various cellular mechanisms, we now investigate how GC-pair richness of DNA may influence strand displacement by KF (exo-).Considering the aforementioned advantages of the D-loop structure, we use D-loop for the following studies. We consider two cases: all-GC sequences and non-all-GC sequences.
We start with non-all-GC sequences. Several representative FRET traces are shown in Figs.3(a)–3(c);and the distribution of the starting value of the ascending FRET jumps is given in Fig.3(d),which shows that KF backsliding occurs more frequently at FRET values from 0.2 to 0.5. In other words, the backsliding is prone to appear after several base pairs of the hairpin having been unwound. As the sequence of the hairpin DNA used in our smFRET assay starts with GTTCGTCG,which has more GC pairs in the rear part,this result indicates that GC-richness helps to increase the backsliding probability.
We then investigate the backsliding phenomenon in the case that the hairpin DNA is composed of all GC sequences.As qualitatively shown in Figs.4(a)–4(c),the backward steps now appear more frequently and can occur at various FRET values; as clearly shown in Fig. 4(d), which gives the distribution of the starting value of the ascending FRET jumps, a large fraction of the observed backward steps occur at high FRET values (>0.5), indicating that backsliding may occur right after the binding of KF.Compared with the results given in Fig.3,these results for all-GC sequences confirm that backsliding of KF can be facilitated by the GC-richness of DNA sequences.
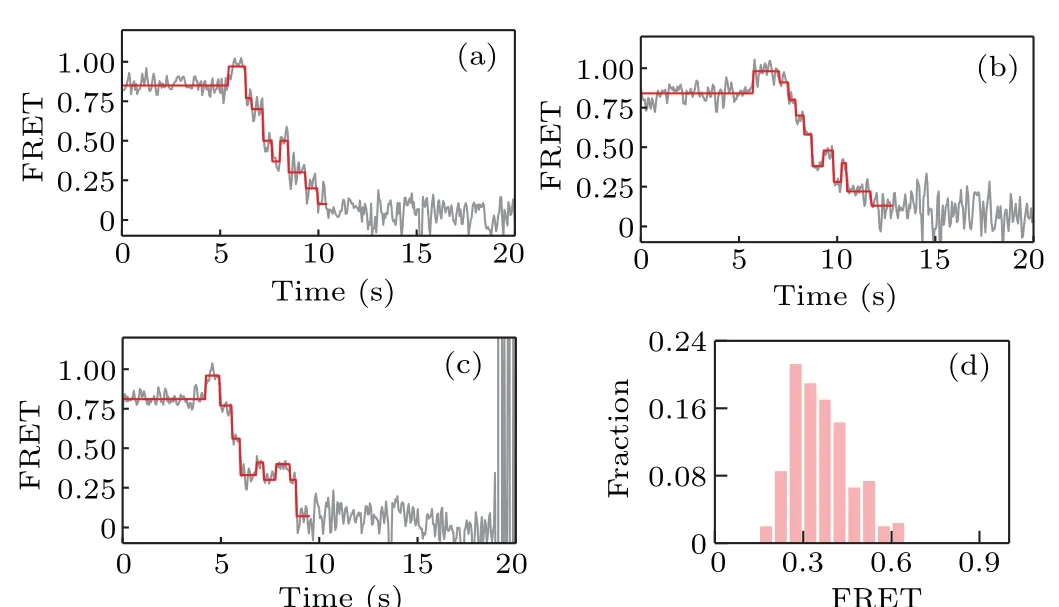
Fig.3. (a)–(c)Representative FRET traces for strand displacement on nonall-GC sequences of D-loop DNA; (d) distribution of the starting value of the ascending FRET jumps.
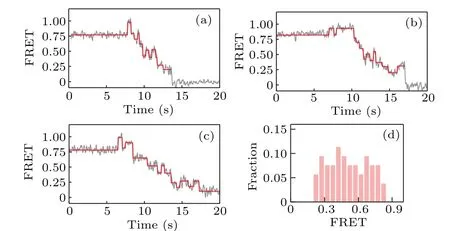
Fig.4. (a)–(c)Representative FRET traces and(d)distribution of the starting value of the ascending FRET jumps, for strand displacement in all-GC sequences.
3.4. Influence of dNTP concentration on strand displacement
We now proceed to investigate how the concentration of dNTP influences the strand displacement phenomenon. At first we use 1-µM concentration of dNTP,and then we change the concentration of dNTP to 10µM and 100µM,respectively.The corresponding representative traces of the FRET value are shown in Fig. 5. When compared to the FRET trace from 1 µM (Fig. 5(a)), those from 10 µM (Fig. 5(b)) and 100 µM(Fig.5(c))seem to have less backward steps. In another word,increasing the dNTP concentration facilitates the unwinding of DNA and thus reduces the possibility of KF backsliding.
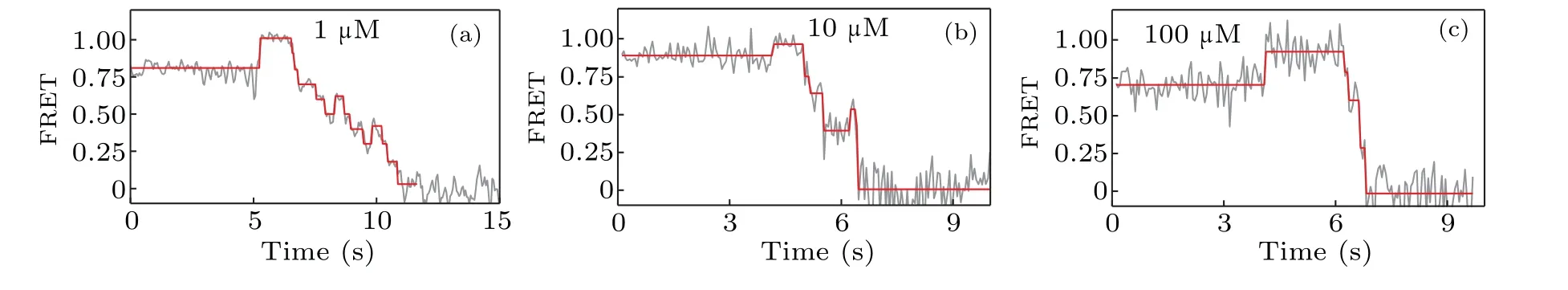
Fig. 5. The dNTP concentration dependence of the strand displacement: (a) typical FRET trace at 1-µM concentration of dNTP, (b) typical FRET trace at 10µM,and(c)typical FRET trace at 100µM.
4. Discussion and conclusion
smFRET experiments on DNA pols were designed to measure DNA polymerization rate by strand displacement of polymerases.[23–25]It was previously reported that 85% of pauses in the strand displacement were sequencedependent and located in the GC-rich region of pol I(Klenow fragment).[23]Our results show that the pauses in strand displacement are related to the backsliding of Klenow fragment on DNA, rather than the stalling of Klenow fragment. As GC-rich sequences are more stable due to base stacking, the presence of GC pairs thus enhances the energy barrier of unwinding and accordingly increases the probability of Klenow fragment backsliding. Our results also suggest that the probability of backsliding decreases as the concentration of dNTP increases. Further studies are needed to elucidate the detailed mechanism behind the influences of GC-richness and dNTP concentration on the backsliding of Klenow fragment.
Acknowledgements
Project supported by the National Natural Science Foundation of China (Grant No. 12090051), the CAS Key Research Program of Frontier Sciences (Grant Nos. QYZDJSSW-SYS014 and ZDBS-LY-SLH015), and the Youth Innovation Promotion Association of CAS(Grant No.2017015).
猜你喜欢
杂志排行
Chinese Physics B的其它文章
- Direct measurement of two-qubit phononic entangled states via optomechanical interactions
- Inertial focusing and rotating characteristics of elliptical and rectangular particle pairs in channel flow
- Achieving ultracold Bose–Fermi mixture of 87Rb and 40K with dual dark magnetic-optical-trap
- New experimental measurement of natSe(n,γ)cross section between 1 eV to 1 keV at the CSNS Back-n facility
- Oscillation properties of matter–wave bright solitons in harmonic potentials
- Synchronously scrambled diffuse image encryption method based on a new cosine chaotic map
