RGB1 Regulates Rice Panicle Architecture and Grain Filling Through Monitoring Cytokinin Level in Inflorescence Meristem and Grain Abscisic Acid Level During Filling Stage
2021-07-13ZhangDongpingZhangMinyanWangYuzhuLiangJiansheng
Zhang Dongping, Zhang Minyan, Wang Yuzhu, Liang Jiansheng
Letter
Regulates Rice Panicle Architecture and Grain Filling Through Monitoring Cytokinin Level in Inflorescence Meristem and Grain Abscisic Acid Level During Filling Stage
Zhang Dongping1, 2, Zhang Minyan1, Wang Yuzhu1, 2, Liang Jiansheng2
(Key Laboratory of Molecular Design for Plant Cell Factory of Guangdong Higher Education Institute, Department of Biology, Southern University of Science and Technology, Shenzhen 518055, China; Jiangsu Key Laboratory of Crop Genetics and Physiology / Co-Innovation Center for Modern Production Technology of Grain Crops, Key Laboratory of Plant Functional Genomics of the Ministry of Education, Yangzhou University, Yangzhou 225009, China)
regulated not only rice panicle architecture but also the grain filling process. Suppression ofexpression resulted in a denser panicle and smaller grain size and weight, which was largely due to the lower endogenous cytokinin (CK) level caused by the increased expression of cytokinin oxidase genes in the inflorescence meristem. Suppression ofexpression also significantly delayed the grain development and reduced the starch accumulation rate through reducing grain abscisic acid (ABA) accumulation during early filling stage. Exogenous ABA can partially rescue the detrimental effects of suppression ofexpression on starch accumulation and grain weight. In conclusion,regulated rice panicle architecture and grain filling through monitoring CK level in the inflorescence meristem and grain ABA level during filling stage.
Rice yield is, to a large extent, determined by the number of spikelets borne on the panicles per unit area and the grain weight (Li et al, 2020). The number and the pattern of panicle branches determine the number and weight of grains on them, which is a main factor affecting the grain yield. Therefore, understanding the regulatory mechanisms underlying the formation of branches and the grain filling is critical to devise strategies for breeding programs.
In the past decades, a number of rice varieties with large panicle bearing numerous spikelets have been bred in China. The higher grain yield in super hybrid rice cultivars is attributed to larger panicle size coupled with higher biomass production or higher harvest index. However, the factors controlling the differentiation of the panicle meristem, and thus the size of panicle and the number of spikelets on it are largely unknown. Several QTLs/genes that determine the panicle size and the number of spikelets have been identified and cloned (Li et al, 2016). One of the most effective QTLs cloned for panicle architecture is, which controls panicle size (Huang et al, 2009; Zhou et al, 2009). The effect of dominantallele is to enhance meristematic activity, resulting in a reduced length of the inflorescence internode, an increased number of grains per panicle and a consequent increase in grain yield (Huang et al, 2009; Zhou et al, 2009). A single nucleotide polymorphism (G/C) at the -1 253 bp of the promoter region ofcauses a core sequence shift (TGGGCC) of a site II transcriptional regulatory element and largely affects the numbers of primary and secondary branches as well as number of grains per panicle (Zhao et al, 2016). Another gene,(erect-pose panicle), markedly enhances grain yield, chiefly by increasing number of secondary branches and number of grains on the secondary branch.gene also produces a remarkable increase in grain density (Wang et al, 2009). However, the mechanisms of these two genes in regulating rice panicle architecture and grain number are largely unknown. Liu et al (2013) reported that the genetic pathway determining inflorescence architecture is conserved betweenand rice, and four rice orthologs of MADS-box transcription factors, are also involved in the regulation of panicle branching through regulating-like genes in rice. The pioneering work of the molecular analysis of grain number determination in rice is the identification of the gene() (Ashikari et al, 2005).encodes cytokinin oxidase/ dehydrogenase (OsCKX2), which degrades the phytohormone CK. The reduced expression ofcauses CK accumulation in the inflorescence meristem and increases the number of reproductive organs, resulting in enhanced grain number. Either the loss of function mutation ofor the mutation of the gene using CRISPR/Cas9 editing technique results in enhanced grain number per panicle (Li et al, 2016).
The larger panicle varieties often show larger sink size and higher yield potential over conventional varieties (Laza et al, 2004; Jiang et al, 2014). However, the majority of these large panicle varieties have always failed to achieve the expected yield performance in production, mainly due to poor grain filling (Zhang et al, 2012; Li et al, 2013). A lot of studies have been conducted to explore the causes of poor grain filling in the large panicle rice varieties, but the physiological and molecular mechanisms underlying controlling grain filling are still largely elusive. Within the panicle, individual grains at different positions/branches develop sequentially and are at different development stages. Usually, the florets on the upper central axis, the upper branches and the primary branches develop first (earlier flowering and pollination) and are called superior grains (SGs), while the florets on the lower branches, the secondary and tertiary branches are called inferior grains (IGs). Great differences have been observed between SGs and IGs in the growth and developmental characteristics at the levels of cell biology, physiological and biochemistry, even at the level of molecular biology, especially for those large panicle rice varieties (Tang et al, 2009; Yang and Zhang, 2010; Peng et al, 2011; Zhu et al, 2011; You et al, 2016, 2017; Wang et al, 2019).
Information retrieved from the public microarray database show that high levels oftranscripts are found during panicle development (Jain et al, 2007). To investigate whetherhas function in the regulation of panicle architecture and number of grains per panicle in rice, twoRNA-interfered transgenic lines (and) were generated. Compared with the wild type, Wuyujing 8 (WYJ8),lines had much shorter panicle length and fewer grain numbers (Fig. 1-A to -C), suggesting thatwas involved in the regulation of both panicle length and grain number per panicle. Furthermore,lines also had a smaller grain size and lower 1000-grain weight (Fig. S1), which is very similar to the results of Utsunomiya et al (2011) and Sun et al (2018). These results suggested thatcontrolled the panicle development and grain formation as well as the grain size and weight. However, its mechanisms remain elusive.
Reduced expression ofcauses CK accumulation in inflorescence meristems and increases the number of grains per panicle (Ashikari et al, 2005), implying that CK plays a key role in controlling grain formation.In this study, we measured the contents of zeatin (Z) and zeatin-riboside (ZR) in the inflorescence meristem of different genotypes. The endogenous levels of Z and ZR in the inflorescence meristem were substantially higher in WYJ8 than in the two transgenic rice lines with suppressed expression of(Fig. 2-A), which implied thatfunction in the regulation of panicle architecture and grain number might relate to the changes of endogenous CK level in the inflorescence meristem. However, the mechanisms underlying howcontrols the endogenous CK level in the inflorescence meristem are still unknown.
Endogenous CK level in the inflorescence meristem is determined by its biosynthesis and catabolism, possibly also by the CK transport between cells. Here, the expression of CK oxidase/dehydrogenase (CKX) genes in the inflorescence meristem was quantitatively analyzed. In rice genome, there are totally 11 genes encoding CKXs, however, only 7 genes expressed in the inflorescence meristem (Fig. 2-B). The transcript levels of five CKX genes (,,,and) were significantly higher inlines than those in WYJ8 (Fig. 2-C), whereasandwere much lower as compared with those in(Fig. S2). Becauseandwere much highly expressed in the inflorescence meristem of rice, it was reasonable to assume thatpositively regulated endogenous CK levels largely through inhibiting the expression ofand, although its possible regulation on the expression of genes encoding proteins/enzymes related to CK biosynthesis and transport between cells can not be ruled out.

Fig. 1. Comparisons of panicle phenotype, grain filling characteristics and effects of abscisic acid (ABA) application between Wuyujing 8 (WYJ8) andtransgenic lines.
A, Panicle phenotype. Scale bar, 1.5 cm. B, Panicle length (= 10). C, Number of grains per panicle (= 10). D, Increase in grain dry weight after flowering (= 10). E and F, Grain starch content and starch accumulation rate at the different days after flowering and the effects of ABA application. Values are Mean ± SD (= 3). * and ** indicate significant differences at the 0.05 and 0.01 levels, respectively.
was involved in regulating grain filling process. The grain filling rate increased rapidly and peaked at 10 d after flowering (DAF), and then decreased rapidly till the end of grain filling in WYJ8. However, the grain filling rate oflines increased much slowly at the early stage of grain development and the maximal grain filling rate appeared at 25 DAF. Furthermore, the peak value of the increase in grain dry weight was also much lower inlines (Fig. 1-D to -F), suggesting that suppression ofexpression dramatically delayed the rapid starch accumulation and, as a consequence, reduced the final grain weight (Fig. S1-C).
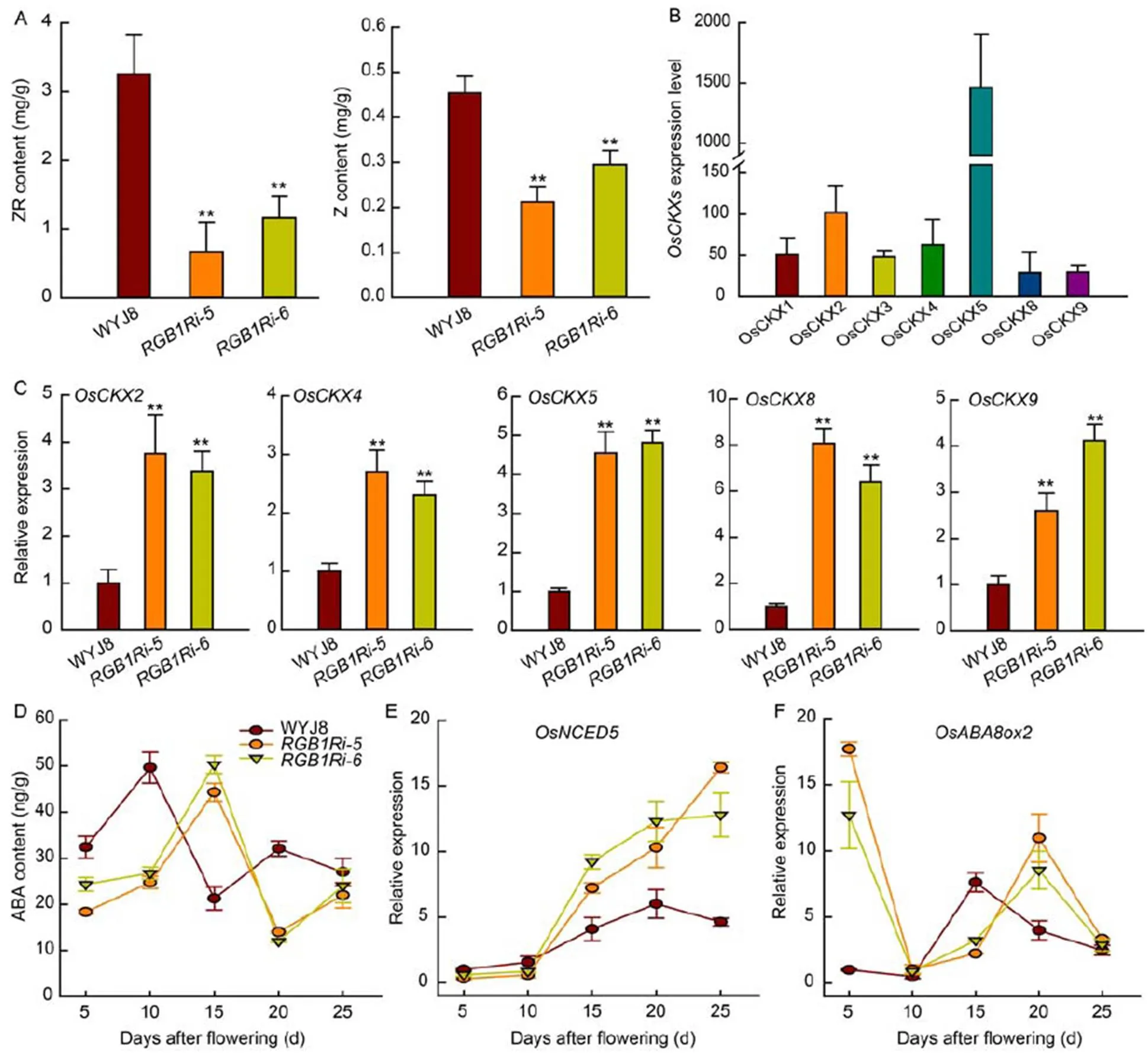
Fig. 2. Contents of cytokinins and abscisic acid (ABA) and expression levels of their related genes.
A, Contents of zeatin-riboside (ZR) and zeatin (Z) in the inflorescence meristem quantitatively analyzed by high-performance liquid chromatography- tandem mass spectrometry (HPLC-MS/MS). B and C, Expression of cytokinin oxidase/dehydrogenase (CKXs) genes in the inflorescence meristem. D, Grain ABA content during grain filling stage quantitatively analyzed by HPLC-MS/MS. E and F, Changes in the expression profiles of ABA synthesis geneand catabolism genein the grains during grain filling stage. Values are Mean ± SD (= 3). ** indicate significant differences at the 0.01 level.
ABA plays important roles in regulating grain filling process (Liang et al, 2001; Tang et al, 2009; Wang et al, 2015). However, whether and how ABA is involved in theregulation of grain filling process is still unknown. In this study, we measured the grain ABA contents during grain filling stage of WYJ8 andtransgenic lines. The results showed that ABA content in WYJ8 grains increased rapidly after flowering and peaked at 10 DAF and decreased thereafter, whereas the grain ABA content intransgenic lines was lower and increased slowly at the early stage of grain filling. The peak appeared at the 15 DAF (Fig. 2-D). The timing coincided with the starch accumulation and grain filling rate (Fig. 1-D). These results suggested that delay and reduction of grain starch accumulation might be related to the low grain ABA content in knock-down lines.
To verify this hypothesis, exogenous ABA was applied on the developing grains during grain filling stage, and starch accumulation and expression of key genes encoding starch biosynthesis were analyzed. ABA application had no obvious effects on both starch accumulation and grain filling rate in WYJ8 grains, but significantly enhanced starch accumulation and accelerated the increase of starch content inlines at the early stage of grain filling (Fig. 1-E and -F). We further investigated the effects of ABA application on the expression of genes encoding enzymes related to starch biosynthesis. Applications of ABA significantly stimulated the expression of eight starch biosynthesis genes (,,,,,,and) in grains of both WYJ8andline at 5 DAF (Fig. S3). Therefore, ABA played important roles in regulating starch biosynthesis andexerted its effect on grain development and starch biosynthesis, at least in part, through regulating grain ABA homeostasis.
During rice grain filling process, ABA content in developing grains fluctuates continuously, which is regulated by multi-stage processes involving ABA synthesis, catabolism and transport, and also affected by environmental conditions exposed. The expression of ABA-signaling and ABA-synthesis genes significantly increase after 3 DAF (Zhang et al, 2020). Studies on the ABA content of developing rice grains showed an increase of ABA from 4 to 14 d after pollination (Wang et al, 2015; You et al, 2016).ABA biosynthesis might be responsible for the ABA accumulation in the endosperm cells (Wang et al, 2015). However, the relative expression of,a key gene encoding the rate-limiting enzyme 9--epoxycarotenoid dioxygenase (NCED) in the endosperm cells, was low at the early stage of grain development, although the expression ofin the WYJ8 grains was higher, but not significant, than that intransgenic lines (Fig. 2-E). Therefore, ABA biosynthetic pathway might contribute little to the increase of ABA content in grains at the early stage of grain development (Fig. 2-D). Understandably, the cellular ABA homeostasis was strictly coordinated in the fashion of complex networks, and the endogenous level of ABA was regulated not only by its biosynthesis, but also by its catabolism. A much higher expression ofin the grains oftransgenic lines was observed at 5 DAF and the expression level decreased rapidly thereafter to the lowest level at 10 DAF. Comparatively, the expression ofin the grains of WYJ8 kept a very low level at the early stage of grain filling (before 10 DAF) (Fig. 2-F). These results indicated that the lower activity of ABA catabolism, rather than ABA biosynthesis, was the main pathway responsible for the elevated ABA accumulation in grains and-modulated ABA homeostasis in grains might be through regulating the expression of.
Taken together, RGB1, a β-subunit of heterotrimic G protein, plays important roles in controlling both the panicle development and the grain filling process in rice.positively controlled the panicle architecture and grain number per panicle through down-regulating the expression of OsCKXs and thus increasing CK level in the inflorescence meristem. Knock-down ofdelayed grain-filling process, and finally reduced grain starch content, at least in part, by delaying an increase of ABA in the endosperm cells, which was caused by higher expression ofat the early stage of grain filling.
ACKNOWLEDGEMENTs
This study was supported by the Peacock Science and Technology Innovation Project of Shenzhen, China (Grant No. KQJSCX20170328153821275), the National Natural Science Foundation of China (Grant No. 32070292), Shenzhen Government Fund for Fundamental Research (Grant No. JCYJ20170817104 523456) and Shenzhen Science and Technology Program (Grant No. KQTD20190929173906742) in China.
SUPPLEMENTAL DATA
The following materials are available in the online version of this article at http://www.sciencedirect.com/journal/rice-science; http://www.ricescience.org.
File S1. Methods.
Fig. S1. Comparison of grain size and 1000-grain weight between different genotypes.
Fig. S2. Expression level ofandin Wuyujing 8 andtransgenic lines.
Fig. S3. Effects of abscisic acid application on expression of genes catalyzing sucrose catabolism and starch biosynthesis.
Ashikari M, Sakakibara H, Lin S Y, Yamamoto T, Takashi T, Nishimura A, Angeles E R, Qian Q, Kitano H, Matsuoka M. 2005. Cytokinin oxidase regulates rice grain production., 309: 741–745.
Huang X Z, Qian Q, Liu Z B, Sun H Y, He S Y, Luo D, Xia G M, Chu C C, Li J Y, Fu X D. 2009. Natural variation at thelocus enhances grain yield in rice.,41: 494–497.
Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP. 2007. F-box proteins in rice: Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress.,143(4): 1467–1483.
Jiang C M, Cheng Z Q, Zhang C, Yu T Q, Zhong Q F, Shen J Q, Huang X Q. 2014. Proteomic analysis of seed storage proteins in wild rice species of thegenus., 12(1): 51.
Laza M R C, Peng S B, Akita S, Saka H. 2004. Effect of panicle size on grain yield of IRRI-releasedrice cultivars in the wet season., 7: 271–276.
Li M R, Li X X, Zhou Z J, Wu P Z, Fang M C, Pan X P, Lin Q P, Luo W B, Wu G J, Li H Q. 2016. Reassessment of the four yield-related genes,,, andin rice using a CRISPR/Cas9 system.,7: 377.
Li P P, Zhu YJ, Guo L, Zhuang JY, Fan YY. Fine mapping of, a minor QTL for grain length, using near isogenic lines derived from residual heterozygotes in rice., 2020, 34(2): 125–134. (in Chinese with English abstract)
Li S Y, Zhao B R, Yuan D Y, Duan M J, Qian Q, Tang L, Wang B, Liu X Q, Zhang J, Wang J, Sun J Q, Liu Z, Feng Y Q, Yuan L P, Li C Y. 2013. Rice zinc finger protein DST enhances grain production through controllingexpression., 110(8): 3167–3172.
Liang J S, Zhang J H, Cao X Z. 2001. Grain sink strength may be related to the poor grain filling ofrice () hybrids.,112(4): 470–477.
Liu C, Teo W Z N, Bi Y, Song S Y, Xi W Y, Yang X B, Yin Z C, Yu H. 2013. A conserved genetic pathway determines inflorescence architecture inand rice., 24(6): 612–622.
Peng T, Lv Q, Zhang J, Li J Z, Du Y X, Zhao Q Z. 2011. Differential expression of the microRNAs in superior and inferior spikelets in rice ()., 62(14): 4943–4954.
Sun S Y, Wang L, Mao H L, Shao L, Li X H, Xiao J H, Ouyang Y D, Zhang Q F. 2018. A G-protein pathway determines grain size in rice.,9: 851.
Tang T, Xie H, Wang Y X, Lü B, Liang J S. 2009. The effect of sucrose and abscisic acid interaction on sucrose synthase and its relationship to grain filling of rice (L.).,60(9): 2641–2652.
Utsunomiya Y, Samejima C, Takayanagi Y, Izawa Y, Yoshida T, Sawada Y, Fujisawa Y, Kato H, Iwasaki Y. 2011. Suppression of the rice heterotrimeric G protein β-subunit gene,, causes dwarfism and browning of internodes and lamina joint regions.,67(5): 907–916.
Wang G Q, Li H X, Feng L, Chen M X, Meng S, Ye N H, Zhang J H. 2019. Transcriptomic analysis of grain filling in rice inferior grains under moderate soil drying., 70(5): 1597–1611.
Wang J Y, Nakazaki T, Chen S Q, Chen W F, Saito H, Tsukiyama T, Okumoto Y, Xu Z J, Tanisaka T. 2009. Identification and characterization of the erect-pose panicle geneconferring high grain yield in rice (L.)., 119: 85–91.
Wang Z Q, Xu Y J, Chen T T, Zhang H, Yang J C, Zhang J H. 2015. Abscisic acid and the key enzymes and genes in sucrose- to-starch conversion in rice spikelets in response to soil drying during grain filling., 241: 1091–1107.
YangJ C, Zhang J H. 2010. Grain-filling problem in ‘super’ rice.,61(1): 1–5.
You C C, Zhu H L, Xu B B, Huang W X, Wang S H, Ding Y F, Liu Z H, Li G H, Chen L, Ding C Q, Tang S. 2016. Effect of removing superior spikelets on grain filling of inferior spikelets in rice., 7: 1161.
You C C, Chen L, He H B, Wu L Q, Wang S H, Ding Y F, Ma C X. 2017. iTRAQ-based proteome profile analysis of superior and inferior spikelets at early grain filling stage inrice., 17: 100.
Zhang D P, Zhang M Y, Zhou Y, Wang Y Z, Shen J Y, Chen H Y, Zhang L, Lü B, Liang G H, Liang J S. 2019. The rice G protein γ subunit DEP1/qPE9-1 positively regulates grain-filling process by increasing auxin and cytokinin content in rice grains., 12(1): 91.
Zhang H, Li H W, Yuan L M, Wang Z Q, Yang J C, Zhang J H. 2012. Post-anthesis alternate wetting and moderate soil drying enhances activities of key enzymes in sucrose-to-starch conversion in inferior spikelets of rice., 63(1): 215–227.
Zhang X F, Tong JH, BaiAN, LiCM, XiaoLT, Xue H W. 2020. Phytohormone dynamics in developing endosperm influence rice grain shape and quality., 62(10): 1625–1637.
Zhao M Z, Sun J, Xiao Z Q, Cheng F, Xu H, Tang L, Chen W F, Xu Z J, Xu Q. 2016. Variations in() contribute to the diversity of the panicle trait in high-yieldingrice varieties in northern China., 66(4): 599–605.
Zhou Y, Zhu J Y, Li Z Y, Yi C D, Liu J, Zhang H G, Tang S Z, Gu M H, Liang G H. 2009. Deletion in a quantitative trait geneassociated with panicle erectness improves plant architecture during rice domestication., 183(1): 315–324.
Zhu G H, Ye N H, Yang J C, Peng X X, Zhang J H. 2011. Regulation of expression of starch synthesis genes by ethylene and ABA in relation to the development of rice inferior and superior spikelets., 62(11): 3907–3916.
Liang Jiansheng (liangjs@sustech.edu.cn)
18 August 2020;
12 January 2021
Copyright © 2021, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2021.05.002
杂志排行
Rice Science的其它文章
- SDF5 Encoding P450 Protein Is Required for Internode Elongation in Rice
- Application of Rice Husk Biochar for Achieving Sustainable Agriculture and Environment
- MULTI-FLORET SPIKELET 4 (MFS4) Regulates Spikelet Development and Grain Size in Rice
- OsbZIP09, a Unique OsbZIP Transcription Factor of Rice, Promotes Rather Than Suppresses Seed Germination by Attenuating Abscisic Acid Pathway
- A New Approach to Select Doubled Haploid Rice Lines under Salinity Stress Using Indirect Selection Index
- iTRAQ-Based Proteomics Investigation of Critical Response Proteins in Embryo and Coleoptile During Rice Anaerobic Germination
