含季铵盐的芳酰腙配体的铜 (Ⅱ)配合物的合成和表征:体外DNA键合和核酸酶活性
2020-07-20EdaengElifCansuTopkayaTolgaktrkRamazanGup
Eda Şengül Elif Cansu Gökçe Topkaya Tolga Göktürk Ramazan Gup
(Department of Chemistry,Faculty of Science,Mugla Sıtkı Koçman University,48000,Kotekli-Mugla,Turkey)
0 Introduction
DNA is the storage and transport of genetic information in the cell,and it is a prime target molecule for the drugs.The interaction of drugs with DNA can cause DNA damage in cancer cells,blocking the division of cancer cells and resulting in cell death[1-2].After success of cisplatin as an anticancer agent the design and use of metal based compounds as antitumor drugs have been become one of the most interdisciplinary research area for medicinal inorganic chemists[3-5].On the other hand,the use of platinum metal-based cisplatin drug in cancer therapy causes numerous side effects which unfortunately have limited its use[4]and paved the way for the development of new compounds[6-10].
Inorganic chemistry presents a broad possibility for the design of new metal-based drugs based on the coordination and redox properties of metal complexes to treat cancer[11-15].Currently,various metal complexes are regarded as the most capable replacement for classical cisplatin-type drugs[16-19].Among the various metal complexes,copper-based metal complexes have been extensively investigated due to their low side effects and high biological activities after coordinated with ligands[20-22].
Because of extensively using as chelating ligands in field of coordination chemistry,Schiff bases have been taken notice significant compounds in medicinal chemistry for decades.They can be easily synthesized and coordinated with different kinds of metal ions.Schiff base hydrazone derivatives as well as their coordination compounds have been used for various essential biological activities including anticancer,anti HIV,antibacterial activities,enzyme inhibitors,etc[16-17,23-27].In addition to these,the copper (Ⅱ)complexes of hydrazone ligands show good activity in binding and cleaving DNA[28-31].Therefore,they are one of the important candidates in metal-based drug medicinal researches.
The compounds having quaternary ammonium salts moiety are widely used as bioactive agents.Quaternary ammonium salts have a significant role in the living organisms and are commonly used for the control of bacterial growth in clinical and industrial environments[32-34].Quaternary ammonium salts are used as fungicides,bactericides,antiseptics and therapeutic agents due to their excellent antimicrobial activity.The presence of alkyl chains in drugs increases the lipophilicity properties of compounds and enables these compounds to pass through bio-molecules.Keeping in minds the potential biological activities of hydrazones and quaternary ammonium salts,we have decided to design new copper (Ⅱ)complexes of Schiff base ligands bearing quaternary alkyl ammonium salts and hydrazones.DNA bindingin vitroand DNA cleavage studies have also been evaluated.
1 Experimental
1.1 Materials and instrumentation
All reagents used in this work were purchased from Merck and Sigma-Aldrich and used as supplied.pBR322 DNA was purchased from Fermentas.Microanalysis(C,H,N)were performed on a LECO 932 CHNS analyzer and copper content was measured by atomic absorption spectroscopy using the DV 2000 Perkin Elber ICP-AAS.Magnetic susceptibility measurements were carried out by using a Sherwood Scientific MK1 Model Gouy Magnetic Susceptibility Balance at room temperature.NMR spectra were recorded on a Bruker 400 MHz spectrometer in DMSO-d6with TMS as the internal standard.IR spectra were recorded on pure solid samples with a Thermo-Scientific,Nicolet iS10-ATR.The electronic spectra of the ligands and complexes were recorded on a PG Instruments T80+UV/Vis Spectrophotometer.4-Hydroxybenzohydrazide(Scheme 1)was synthesized by the refluxing of ethyl 4-hydroxybenzoate with hydrazine hydrate for 4 h in ethanol.
1.2 Preparation of 3-(4-acetylphenoxy)-N,N,N-trimethylpropane-1-ammonium perchlorate(A)and 5-(4-acetylphenoxy)-N,N,N-trimethylpentane-1-ammonium perchlorate(B)
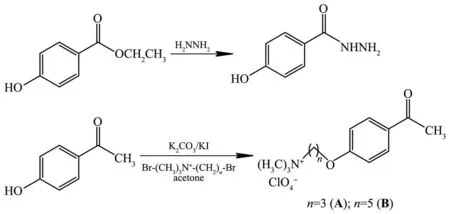
Scheme 1 Schematic diagram showing synthesis of 4-hydroxybenzohydrazide and(4-acetylphenoxy)-N,N,N-trimethylalkyl-1-ammonium perchlorate compounds
K2CO3(1.52 g,0.011 mol)and catalytic amount of KI were added to a solution of 1.36 g(0.01 mol)of 4-hydroxyacetophenone in 20 mL of acetone,followed by stirring for 15 minutes.(3-Bromopropyl)trimethylammonium bromide(0.01 mol,2.87 g)or(5-bromopentyl)trimethylammonium bromide(1.36 g,0.01 mol)was added as a solid and the resulting mixture was stirred under reflux for 24 hours.After evaporating the solvent with evaporator,the remaining solid was dissolved with minimum amount of cold water and an equal amount of NaClO4was added and kept in refrigerator for one night.The resulting white solid precipitate was filtered off.The product was purified by crystallization from methanol.The purity of the compound was checked with thin layer chromatography.
For A:Yield:81%.m.p.150~152 ℃ .UV-Vis(DMF,nm)205.5,216.0,272.0(sh),299.FTIR(ATR,cm−1):2 956(C-H)aliph,1674(C=O),1 246(C-O).1H NMR(DMSO-d6):δ2.19(p,2H,CH2),2.31(s,3H,CH3),3.18(s,9H,N+-CH3),3.45(t,2H,N-CH2),4.09(t,2H,O-CH2),7.75(2H,d,ArH),7.98,(2H,d,ArH).13C NMR(DMSO-d6):δ186.7(C=O),172.3(C=O),162.7(C-O),149.1,147.6 and 142.9(C=N),137.1,131.8,128.4,119.4 and 114.4(Ar-C),64.5(O-CH2),10.4(CH3).Elemental Anal.Calcd.for C14H22ClNO6(%):C,50.08;H,6.60;N,4.17.Found(%):C,50.01;H,6.53;N,4.15.
For B:Yield:79%.m.p.129~131 ℃.UV-Vis(DMF,nm):205.5,271.0,291(sh),304.0.FTIR(ATR,cm−1):2 956(C-H)aliph,1 669(C=O),1 261(C-O).1H NMR(DMSO-d6):δ1.14(2H,p,CH2),1.77(4H,m,CH2),2.45(3H,s,CH3),3.15(9H,s,N-CH3),3.28(2H,t,N-CH2),4.06(2H,t,O-CH2),7.01(2H,d,ArH)7.90(2H,d,ArH).13C NMR(DMSO-d6):197.0(C=O),163.1(C-O),131.2,130.5 and 114.9(Ar-C),68.1(O-CH2),65.8(N-CH2),52.8(N-CH3),28.6(CH3),27.1,23.0 and 22.5(CH2).LC-MS:m/z364.15 for[M]+.Elemental Anal.Calcd.for C16H26ClNO6(%):C,52.82;H,7.20;N,3.85.Found(%):C,52.75;H,7.23;N,3.87.
1.3 Synthesis of the hydrazone Schiff base ligands
A or B(1 mmol)was added as a solid to a solution of 4-hydroxybenzohydrazide(1 mmol)dissolved in 10 mL of ethanol with two drops of glacial acetic acid.The reaction mixture was refluxed for further 24 h.The precipitated compound was collected by filtration,washed with diethyl ether and then dried in vacuum.The product was recrystallized by water-ethanol(2∶1,V/V).
For(E)-3-(4-(1-(2-(4-hydroxybenzoyl)hydrazono)ethyl)phenoxy)-N,N,N-trimethylpropan-1-ammonium perchlorate(H2L1):Yield:69%.m.p.276℃.UV-Vis(DMF,nm):272,306.IR(ATR,cm−1):3 162(OH),2 978(C-Haliph),1 655(C=O)amide,1 609(C=N),1 397(C-N),1 274(C-O),1 101(ClO4−).1H NMR(DMSO-d6):δ2.22(p,2H,CH2),2.33(s,3H,CH3),3.12(s,9H,N+-CH3),3.50(t,2H,N-CH2),4.12(t,2H,O-CH2),6.86(d,2H,J=8.5,ArH),7.02(d,2H,J=8.6,ArH),7.79(d,4H,J=8.6,ArH),10.09(s,1H,OH),10.48(s,1H,NH).13C NMR(DMSO-d6):δ169.4(C=O),160.7(C=N),160.8 and 159.6(ArC-O),130.3,129.2,128.3,124.9,115.2 and 114.4(Ar-C),67.2(CH2-O),63.4(N-CH2),52.7(N+-CH3),23.0(CH3),14.4(CH2).Elemental Anal.Calcd.for C21H28ClN3O7(%):C:53.67,H:6.01,N:8.94.Found(%):C:53.89,H:6.08,N:9.01.
For(E)-3-(4-(1-(2-(4-hydroxybenzoyl)hydrazono)ethyl)phenoxy)-N,N,N-trimethylpentane-1-ammonium perchlorate(H2L2):Yield:72%.m.p.222℃.UV-Vis(DMF,nm):271,309.FTIR(ATR,cm−1):3 391(OH),2 934 and 2 872(C-Haliph),1 650(C=O)amide,1 611(C=N),1 381(C-N),1 257(C-O),1 085(ClO4−).1H NMR(DMSO-d6):δ1.48(p,2H,CH2),1.78(m,4H,CH2),2.32(s,3H,CH3),3.06(s,9H,N+-CH3),3.33(t,2H,N-CH2),4.06(t,2H,O-CH2),6.86(d,2H,J=8.5,ArH),6.99(d,2H,J=8.6,ArH),7.78(d,4H,J=8.6,ArH),10.08(s,1H,OH),10.45(s,1H,NH).13C NMR(DMSO-d6):166.4(C=O),160.(C=N),160.4 and 160.8(ArCO),131.0,129.2,125.0,124.4,115.2 and 114.6(Ar-C),67.6(CH2-O),65.6(N-CH2),52.6(N+-CH3),28.5(CH3),22.8,22.3 and 14.6(CH2).Elemental Anal.Calcd.for C23H32ClN3O7(%):C:55.47,H:6.48,N:8.44.Found(%):C:55.85,H:6.57,N:8.60.
1.4 Synthesis of Cu (Ⅱ)complexes
Copper (Ⅱ)acetate dihydrate(0.20 g,1 mmol)was added as a solid to a solution of H2L1or H2L2(2 mmol)dissolved in MeOH-water(10 mL).Trimethylamine(2 mmol)was added by dropwise to this solution and the reaction mixture was refluxed for 24 h.The precipitated complexes were filtered off and washed with cold water and finally recrystallized by acetonitrile-ethanol(1∶1,V/V).
For[Cu(HL1)2]:Dark brown;Yield:72%;m.p.278 ℃.μeff=1.73μB;UV-Vis(DMF,nm):272.0,308.0,362.0(sh),380.0,400.0(sh).FT-IR(ATR,cm−1):3 368b(O-H),2 928 and 2 821w(C-Haliphatic),1 595m(C=N-N=C),1 370m(C-N),1 243s(C-O),1 090(ClO4−).MS-ES+:m/z999.179 for[M]+.Elemental Anal.Calcd.for C42H54Cl2CuN6O14(%):C:50.38,H:5.44,N:8.39,Cu:6.35.Found(%):C:50.04,H:5.76,N:8.62,Cu:6.10.
For[Cu(HL2)2]:Dark brown;Yield:71%;m.p.277 ℃.μeff=1.70μB;UV-Vis(DMF,nm)274.0,312.0,366.0(sh),384.0 and 402.0(sh).FT-IR(ATR,cm−1):3 423b(O-H),2 951 and 2 874w(C-Haliphatic),1 597m(C=N-N=C),1 366m(C-N),1 247s(C-O),1 100s(ClO4).MSES+:(m/z)1 057.034 for[M]+.Elemental Anal.Calcd.for C46H62Cl2CuN6O14(%):C:52.25,H:5.91,N:7.95,Cu:6.01.Found(%):C:52.48,H:6.04,N:7.78,Cu:6.49.
1.5 DNA binding
1.5.1 Electronic absorption titrations
All the binding experiments of these compounds with CT-DNA were conducted in a buffer containing 5 mmol·L−1Tris(tris(hydroxymethyl)aminomethane)and 50 mmol·L−1NaCl,and adjusted to pH=7.3 with HCl[30,35].The determination of UV-Vis absorption spectra was carried out by adding the increasing amounts of DNA(from 0 to 100 µmol·L−1)to each of these compounds with a fixed concentration(50 µmol·L−1)dissolved in a solvent mixture of 1%(V/V)DMF and 99%(V/V)Tris-HCl buffer.The spectra were measured in a wavelength range of 200~500 nm.
1.6 DNA cleavage
The cleavage of supercoiled pBR322 DNA was determined by agarose gel electrophoresis[35].The gel electrophoresis experiments were performed by incubation of the samples containing 7µL pBR322 plasmid DNA(50 ng· µL−1)and different concentrations of compounds(50,100,150,200 and 250 µmol·L−1)dissolved in DMF in 100 mmol·L−1Tris-HCl buffer(pH 8.0)at 37℃for 2 h in the presence and absence of H2O2(5 µL,5 mmol·L−1).After incubation,the samples were loaded with 4µL loading dye(0.25%bromophenol blue,0.25%xylene cyanol,30%glycerol,10 mmol EDTA)on a 1%agarose gel containing 1 µg·mL−1of EtBr.The gels were run at 100 V for 3 h in TBE buffer and photographed under UV light.To determine binding site and the presence of reactive oxygen species generated during strand scission,reactive oxygen intermediate scavengers(100 µmol·L−1),that is,DMSO,KI,NaN3and catalase or groove binders were added alternately to the reaction mixture.
2 Results and discussion
2.1 Synthesis
The reactions of 4-hydroxyacetophenone with(5-bromopropyl)trimethylammonium bromide and(5-bromopentyl)trimethylammonium bromide in the presence of dry K2CO3,KI and stoichiometric amount of NaClO4gave 3-(4-acetylphenoxy)-N,N,N-trimethylpropane-1-ammonium perchlorate(A)and 5-(4-acetylphenoxy)-N,N,N-trimethylpentane-1-ammonium perchlorate(B),respectively(Scheme 1).The hydrazone Schiff base ligands(H2L1and H2L2)were synthesized by the condensation reaction of compounds A and B with 4-hydroxybenzoic hydrazide in ethanol.The reactions proceeded smoothly producing the corresponding aroylhydrazones in good yields(Scheme 2).They are soluble in common organic solvent but insoluble in water.The structures of the ligands were elucidated by elemental analyses,FTIR,electronic absorption,and1H and13C NMR.The complexes were synthesized by reaction of the hydrazone ligands with copper (Ⅱ)acetate in the molar ratio of 2:1.All the complexes are highly soluble in MeOH,DMF and DMSO.Attempts to isolate crystals suitable for single X-ray diffraction were unsuccessful.Therefore,the new Cu (Ⅱ)complexes were characterized by elemental analysis,mass,IR and UV-visible spectroscopy.The analytical data obtained for the hydrazone Schiff base and the copper (Ⅱ)complexes are well agreement with the proposed molecular formula.

Scheme 2 Synthetic route for the hydrazone Schiff base ligands
2.2 Infrared spectra
The IR spectra of the compounds in a region of 400~4 000 cm−1were analyzed.The IR spectra of compounds A and B gave absorption bands in the region of 1 674~1 669 cm−1corresponding to carbonyl stretching and strong and broad bands at~1 070 cm−1corresponding to the characteristic chlorate ion stretching.The IR spectra of the hydrozone Schiff base ligands H2L1and H2L2showed a new broad O-H stretching vibration at 3 368 and 3 426 cm−1,respectively.The characteristic amide Ⅰ band was observed at 1 655 and 1 650 cm−1while the other characteristic band due to azomethine group appeared at 1 611 and 1 607 cm−1for H2L1and H2L2,respectively.The bands observed in the region of 1 246~1 274 cm−1in all compounds can be assigned toν(C-O)stretching vibration.The strong stretching vibrations observed in the region of 1 674~1 669 cm−1in the IR spectra of free hydrazone ligands disappeared when the hydrazone ligands coordinated to the copper (Ⅱ)ion[15,31].On the other hand,the infrared spectra of the copper complexes exhibited a new band at 1 595 and 1 597 cm−1for[Cu(HL1)2]and[Cu(HL2)2],respectively,due to the asymmetric stretching vibration of the newly formed-N=C<bond,which is probably as a result of the enolization of ligands on coordination[31,36-37].By comparing IR spectra of the copper (Ⅱ)complexes with those of the free ligands,the data give evidence of coordination of the hydrazone ligands to the copper ion via enolate ion and azomethine nitrogen atoms[21,38].The proposed structures of the copper (Ⅱ)complexes were given in the Fig.1.The other peaks observed in the IR spectra of the compounds synthesized in this work are given in experimental section.
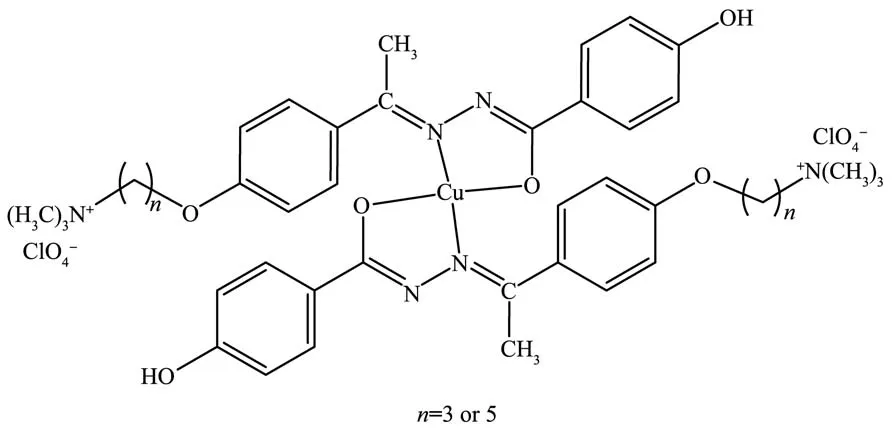
Fig.1 Suggested structures of the copper (Ⅱ)complexes
2.3 NMR spectroscopy
Additional structural information can be deduced from1H NMR and13C NMR spectra for the hydrazine Schiff base ligands.The NMR spectra of the compounds were recorded at room temperature in DMSO-d6.In the1H NMR spectrum of the compounds,the chemical shifts appeared in the region of 3.06~3.18,3.28~3.50 and 4.06~4.12 are attributed to-N+(CH3)3protons,-CH2N protons and-CH2OPh protons,respec-tively.The1H NMR spectra of the compounds exhibited a singlet and a multiple peak in the region of 2.31~2.45 and 1.14~1.17 for the methyl((C=O)-CH3)and the other methylene(CH2)protons,respectively.The chemical shift for the hydroxyl proton(-OH)and-NH proton appeared in the region of 10.08~10.09 and 10.40~10.45,which also indicates the formations of the hydrazone Schiff base ligands.These data are in agreement with that previously reported for similar compounds[38-41].
In the13C NMR spectra of A and B,the chemical shifts at 197.0 and 163.1 are due to carbonyl carbon(C=O)and aromatic C-O,respectively.It is remarkable that these signals of carbonyl carbons(C=O)were replaced by azomethine carbon(C=N)in the hydrazone Schiff base ligands at~160 when the compounds A and B were reacted with 4-hydroxybenzohydrazide.The signals in the region of 131.2~114.9 are ascribed to the carbon atoms of aromatic ring.The signals at 68.1,65.8 and 52.8 are assignable to-O-CH2,-N+-CH2and-N+-(CH3)3carbons,respectively.The chemical shifts corresponding to the aliphatic carbon atoms were observed in the region of 27.1~22.5.In addition,the chemical shifts at 169.4 and 166.4 are attributed to the carbonyl carbon(C=O)of the hydrazide side of H2L1and H2L2ligands,respectively.1H and13C NMR spectra of the copper (Ⅱ)complexes could not be obtained due to their paramagnetic nature.
2.4 Electronic absorption spectra and mass spectra
The electronic absorption bands in the spectra of the ligands and copper (Ⅱ)complexes recorded in dimethylformamide solution are presented in experimental sections.The intense bands in the region of 270~272 nm and 306~309 nm are due toπ→π*andn→π*transitions of the hydrazone ligands,respectively.The shoulders observed in the region of 310~304 nm and 272~274 nm of the copper complexes are attributed ton→πandπ→π*transitions.Then→πtransition,which appeared at 328 nm in the spectrum of the uncoordinated ligands was slightly shifted to a higher wavelength upon coordination.This is an indication of the enolization followed by the deprotonation of the ligand during coordination.The broad bands observed at 380 and 388 nm and the shoulders at 400 and 402 nm are attributed to the intra-ligand and ligand-to-metalcharge transfer(LMCT)transitions,respectively.As expected for the one unpaired electron(S=1/2)system(Cu (Ⅱ)d9),the observed magnetic moment values for[Cu(HL1)2]and [Cu(HL2)2]complexes are 1.73μBand 1.70μB,respectively.
The mass spectra of the copper (Ⅱ)complexes were recorded and represented in Fig.S9 and S10(Supporting information).The mass spectra showed that the molecular peaks observed at[M]+=999.179 and[M]+=1 057.034,are consistent with the molecular weight of complexes[Cu(HL1)2]and[Cu(HL2)2],respectively.
2.5 DNA binding studies
DNA binding is the critical step in the design and construction of new and more efficient drugs targeted to DNA.A compound can bind to the DNA via covalent or non-covalent interactions.Non-covalent binding contains intercalation between the base pairs,binding to major or minor grooves or electrostatic interaction[42-43].
The probable binding ability of the hydrazone ligands and their Cu (Ⅱ)complexes towards CT-DNA were studied by UV-Vis spectroscopy in a range of 250~500 nm.The typical absorption titration curves of the compounds at constant concentration(20 µmol·L−1)in the presence of different concentrations of CTDNA are shown in Fig.2.In the presence of DNA,the absorption bands of the ligands were affected,resulting with considerable hyperchromism with a minor red shift at 270 and 271 nm for H2L1and H2L2,respectively.Similarly,with increase in concentration of CTDNA,the absorption spectra of the Cu (Ⅱ)complexes also exhibited hyperchromism with the minor bathochromic shift(1~2 nm)within the bands at 274 and 272 nm for[Cu(HL1)2]and[Cu(HL2)2],respectively,indicating that all compounds bind with DNA.On the other hand,the bands between 306 and 312 nm for the ligands and their Cu (Ⅱ)complexes show only hypochromism without any shift in wavelength,so that isobestic points are formed in the region of 301~304 nm.These changes are typical of the compounds interacting with DNA via non-covalent interaction[44].Hyperchromism results from the secondary damage of DNA double helix structure[45-46].The hyperchromic effect may also due to the electrostatic interaction between positively(CH3)3N+group of the synthesized compounds and the negatively charged phosphate group of DNA-helix[47-48].
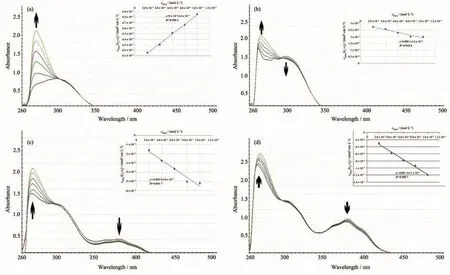
Fig.2 Absorption titration curves of the ligands and their Cu (Ⅱ)complexes in the presence of increasing concentration of CT-DNA
In order to compare quantitatively the DNA binding strength of the ligands and their Cu (Ⅱ)complexes,the intrinsic binding constantsKbof the compounds were determined with Eq.(1)by monitoring the changes in absorbance at~270 nm for the ligands,384 nm for[Cu(HL1)2]and 380 nm for[Cu(HL2)2]with increasing concentration of CT-DNA:
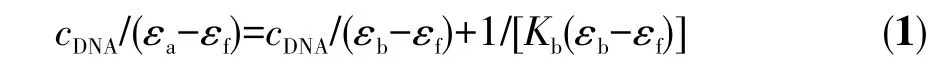
wherecDNAis the concentration of DNA in base pairs,εa,εfandεbcorrespond toAobsd/cM,the extinction coefficient of the complexes and the extinction coefficient of the complex in the fully bound form,respectively,andKbis the intrinsic binding constant.The ratio of the slope to intercept in the plot ofcDNA/(εa−εf)versus cDNAgave the value ofKb.The binding constants obtained for H2L1,H2L2,[Cu(HL1)2]and[Cu(HL2)2]are 3.33×104L·mol−1,1.50×104L·mol−1,1.10×105L·mol−1and 6.25×104L·mol−1,respectively.TheseKbvalues suggest that the H2L1ligand its Cu (Ⅱ)complex have greater binding affinities for calf thymus DNA than the H2L2ligand and its Cu (Ⅱ)complex.
2.7 DNA cleavage studies
Agarose gel electrophoresis has been used for the supercoiled pBR322 DNA cleavage studies of the hydrazone ligands and their copper (Ⅱ)complexes under physiological conditions in the presence or absence of H2O2as an oxidant agent.The DNA cleavage effectiveness was evaluated by determining the ability of these compounds to convert the plasmid DNA from the original supercoiled form(FormⅠ)to the open circular form(FormⅡ)and the linear form(FormⅢ)[49-50].
2.7.1 Hydrolytic cleavage
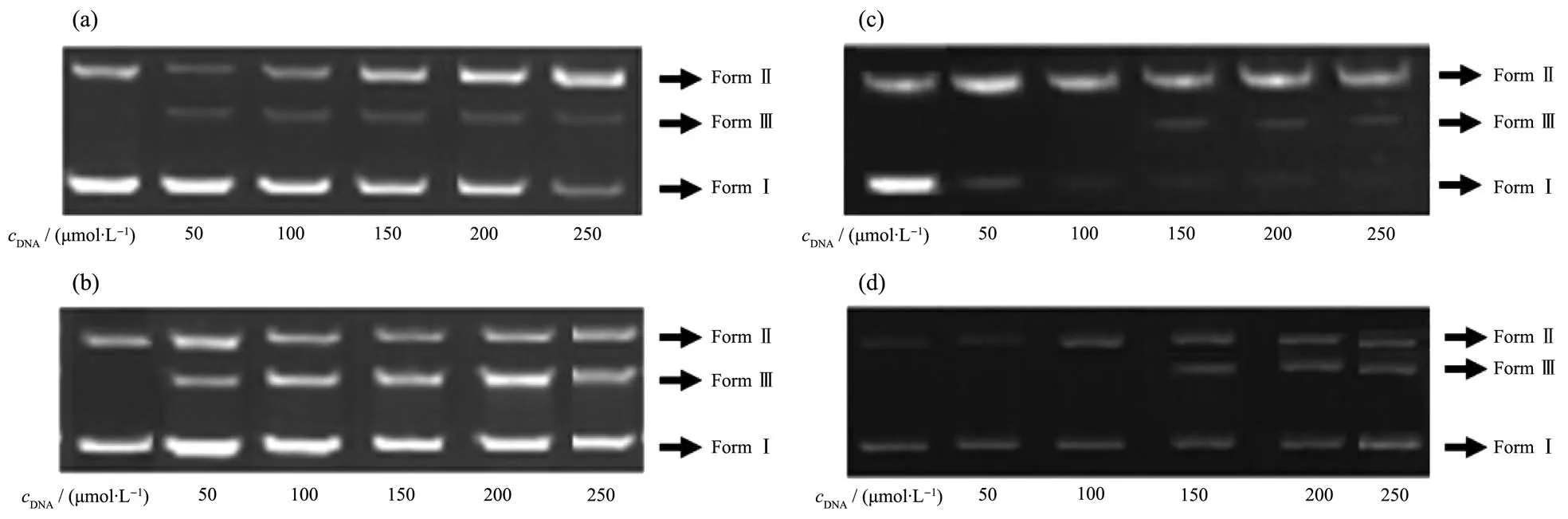
Fig.3 Agarose gel electrophoresis of hydrolytic cleavage of pBR322 DNA mediated by different concentrations of the ligands and their Cu (Ⅱ)complexes after 4 h of incubation
The cleavage activities of the compounds were investigated with different concentrations of the compound in the absence of a co-reagent.With increasing concentration both ligands and their Cu (Ⅱ)complexes have been found to exhibit effective DNA cleavage activity converting the plasmid supercoiled DNA into its cleavage forms(FormⅡ andⅢ)as shown in Fig.3.The ligands cleavaged the supercoiled DNA to nicked and linear DNA at the same time and the increasing intensities of FormsⅡandⅢwere found with the increase of ligand concentration,and the gel pattern indicated the double-strand DNA cleavage.In the case of H2L1,at the concentration of 50 µmol·L−1,the plasmid DNA was slightly cleaved and the percentage of nicked and linear increased with increasing ligand concentration,and finally the supercoiled DNA was almost disappeared when the concentration reached to 250 µmol·L−1(Fig.3(a)).On the other hand,the copper (Ⅱ) complexes of the ligands effectively cleaved supercoiled DNA to open and linear form in high concentrations,surprisingly,compared with the ligands.The complexes slightly cleaved the supercoiled DNA only to nicked form at concentrations of 50 and 100 µmol·L−1.On the other hand,when the concentration of the complex reached to 150 µmol·L−1,linear form was also observed besides nicked form.These data clearly demonstrate that the DNA cleavage activity of the copper (Ⅱ)complexes are better than those of their corresponding ligands.
2.7.2 Oxidative cleavage
The oxidative cleavage of the compounds was also investigated in the presence of H2O2as a co-reactant.The hydrazone H2L2ligand converts the supercoiled DNA to nicked and linear DNA,and all three forms of DNA were observed even at the concentration of 50µmol·L−1(Fig.4(a)and(b)).On the other hand,at 50µmol·L−1,the hydrazone H2L1ligand slightly cleavaged SC DNA into NC and LC forms and the increasing intensities of FormⅡwas found with increase in concen-tration of the H2L1ligand.At 250 µmol·L−1,it was observed that the SC DNA was substantially converted into FormⅡNC and LC forms.

Fig.4 Agarose gel electrophoresis of oxidative cleavage of pBR322 DNA mediated by different concentrations of the ligands and their Cu (Ⅱ)complexes after incubation for 3 h
In comparison with those of in the absence of a coreagent,it is observed that the copper (Ⅱ)complexes enhanced the nuclease activities in the presence of H2O2,which strongly depended on the concentration of the complex.The Cu (Ⅱ)complexes slightly cleaved the supercoiled DNA to FormⅡat concentrations of 50 and 100 µmol·L−1(Fig.4(c)and(d)).On the other hand,linear form also began to appear besides nicked form at the concentration of 150 µmol·L−1.However,the supercoiled form was completely converted into degraded form catalyzed by[Cu(HL1)2]at a concentration of 250µmol·L−1((Fig.4(c)).From these data it can be concluded that the copper (Ⅱ)complexes show better nuclease activity than the ligands in the presence of H2O2.
2.7.3 Mechanism of DNA cleavage
In order to determine whether reactive oxygen species is responsible for the DNA scission process for the compounds,DNA-cleavage reactions mediated by the ligands and their Cu (Ⅱ)complexes were carried out with different inhibiting agents including diffusible hydroxyl radical scavenger(DMSO),superoxide scavenger(KI),singlet oxygen scavenger(NaN3)and hydrogen peroxide scavenger(catalase)both in the presence and absence of an oxidant agent.As can be seen in Fig.5 and 6,the cleavage activities of all compounds were not inhibited markedly in the presence of the radical scavengers used in this work,evidencing that these species are not involved in the cleavage process.All compounds exhibited similar cleavage activities both in the presence and absence of H2O2,making clear that H2O2does not have the critical role in the cleavage reaction catalyzed by all compounds.It may be deduced from these results that DNA is probably cleaved by a hydrolytic pathway since the nuclease activity of the complex is not inhibited in presence of radical scavengers either in presence and absence of hydrogen peroxide.
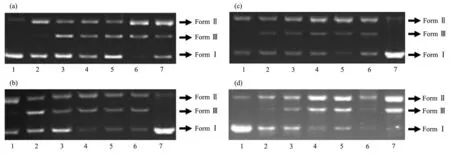
Fig.5 Oxidative cleavage of pBR322 DNA in the presence of different scavengers and groove bindings(methyl green(MG)and 4,6-diamidino-2-phenylindole(DAPI))after 3 h incubation of the ligands and Cu (Ⅱ)complexes
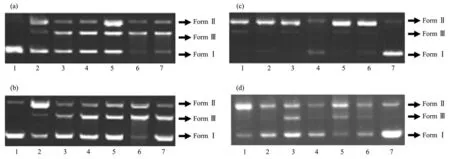
Fig.6 Hydrolytic cleavage of pBR322 DNA in the presence of different scavengers and groove bindings(methyl green(MG)and 4,6-diamidino-2-phenylindole(DAPI))after 4 h incubation of the ligands and Cu (Ⅱ)complexes
To investigate whether the compounds interact with plasmid pBR 322 DNA by groove binding,the cleavage reactions were also carried out in the presence of 4,6-diamidino-2-phenylindole(DAPI)and methyl green,which are known to bind to DNA at minor groove and major groove,respectively,since the UV-visible absorption titration results indicate that the compounds bind to DNA via groove binding mode[51].No apparent inhibition of DNA damage was observed in the presence of DAPI(Fig.5 and 6;lane 2 and 3)neither in the presence nor absence of H2O2indicating that the compounds do not bind to DNA via minor binding.On the other hand,addition of methyl green into the cleavage system relatively inhibited the DNA cleavage for all compounds(Fig.5 and 6,lane 1 and 2)evidencing the major groove preference of both hydrazone ligands and their Cu (Ⅱ)complexes.
3 Conclusions
Herein,two new hydrazone Schiff base ligand bearing quaternary alkyl ammonium salts and their Cu (Ⅱ)complexes have been successfully synthesized and characterized.The new hydrazone Schiff base compounds behave as monobasic bidentate ligands coordinating through azomethine nitrogen and enolic oxygen.All the final compounds have been screened for DNA binding and DNA cleavage.Thein vitroDNA binding studies of both ligands and Cu (Ⅱ)complexes reveal the groove binding,possibly major groove binding mode,which is also supported by the result of the agarose gel electrophorese of DNA scission in the presence of groove binders.The compounds cleave the circular supercoiled DNA to nicked DNA and linear DNA form both in the presence and absence of H2O2,which has strong dependence on the concentration of compounds.Various radical scavengers do not effectively inhibit the DNA cleavage mediated by the titled compounds either in the presence or absence of H2O2,suggesting that the diffusible radicals are not produced during the DNA cleavage by the compounds according to the mechanistic studies.These observations seem to suggest that both hydrazone ligands and their copper (Ⅱ)complexes cleavage the DNA by a hydrolytic pathway.The results also indicate that the chemical nuclease activities of the titled copper (Ⅱ)complexes are higher than those of their corresponding ligands,probably due to their higher DNA binding affinities.
Acknowledgements:This paper has been granted by the Muğla Sıtkı Koçman University Research Projects Coordination Office through Project(Grant No.15/168).We thank to Mugla Sıtkı Koçman University for financial support of this study.
Disclosure statement:No potential conflict of interest was reported by the authors.
Supporting information is available at http://www.wjhxxb.cn
