New record of Japanese snake blenny Xiphasia matsubarai(Perciformes: Blenniidae) from South China Sea*
2018-08-02YIMurong易木荣ZHAOChunxu招春旭SUXin宿鑫TAOYajin陶雅晋YANYunrong颜云榕
YI Murong (易木荣) ZHAO Chunxu (招春旭) SU Xin (宿鑫) TAO Yajin (陶雅晋) YAN Yunrong (颜云榕)
1 College of Fisheries, Guangdong Ocean University, Zhanjiang 524088, China
2 Center of South China Sea Fisheries Resources Monitoring and Assessment, Guangdong Ocean University, Zhanjiang 524088,China
3 Guangdong Provincial Engineering and Technology Research Center of Far Sea Fisheries Management and Fishing of South China Sea, Zhanjiang 524088, China
Abstract Specimens belonging to the family Blenniidae were collected in a fishery resource investigation from the coastal waters of Xisha Islands and Hainan Island, South China Sea in 2016. Combining morphological results with sequence analysis, we identi fied one specimen as Xiphasia matsubarai Okada& Suzuki, 1952. This represents a new record in the South China Sea. In morphology, the specimen has the following traits: body elongated, eel-like or ribbon-like in shape; flanks medium flat; the head small bluntly rounded anteriorly and without a moustache; eyes is slightly smaller, on upper lateral position of head, which is about equal to 1/5 of the length of the head; body without scales, lateral line has been degraded; both sides of the upper and lower jaws with a canine; gill is opening at the top of the pectoral fin base, approximately equal to the length of eye diameter. Dorsal fin XI, 96; pectoral fin 10; anal fin II, 95.Head and body grey-brown, including 26 dark grey-brown bands; abdomen and lower operculum yellowish grey and colour lighter; and dorsal base long with dark grey. Origin of dorsal is located over the anterior margin of pupil; black blotch on dorsal fin between 8th and 10th dorsal spine; anal and caudal fins dark grey, pectoral and ventral fins pale yellow. Sequence analysis of cytochrome oxidase subunit I gene ( COI)strongly supports the identity of the specimen as X. matsubarai.
Key word: new record; Xiphasia matsubarai; Blenniidae; South China Sea; DNA barcoding
1 INTRODUCTION
The family Blenniidae includes 387 species and 57 genera, and most of them are distributed in tropical and subtropical waters in the Paci fic, Indian and Atlantic Oceans (Nelson, 2006; Hastings and Springer, 2009). Eighty two of these species in 31 genera have been recorded in neighboring countries of the South China Sea (Randall and Lim, 2000; Shao et al., 2008), while 72 species in 25 genera are recorded in the Chinese marine territories (Cheng and Zheng, 1987; Shen, 1993; Liu, 2008; Chen and Zhang,2015; Tian, 2016). The genus blenniid Xiphasia includes two species but only Xiphasia setifer Swainson 1839 has been recorded from the South China Sea. One specimen of Xiphasia matsubarai Okada & Suzuki 1952 was collected near the Xisha Islands in April 2016. Morphological characters and comparison of the cytochrome oxidase subunit I gene( COI) with the sequences of GenBank to con firm the correctness of our identi fication.
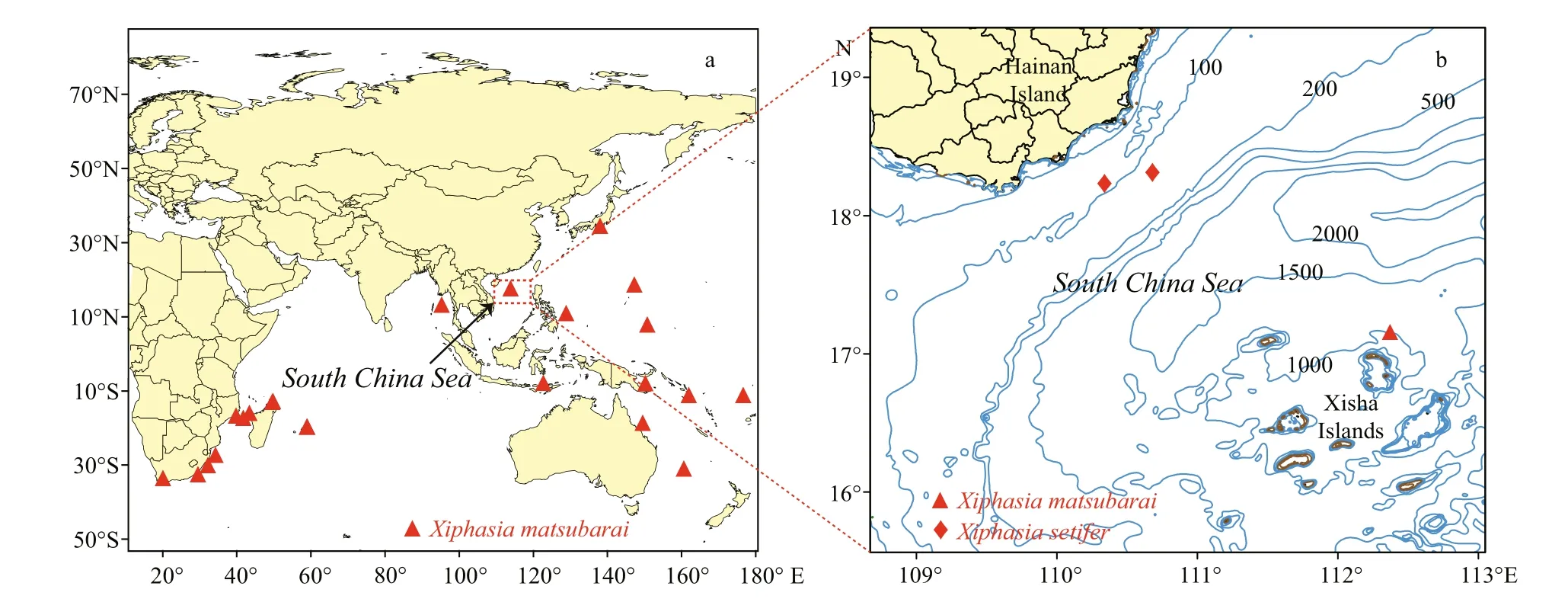
Fig.1 Distributional records for species of X. matsubarai (a) and locations of specimens collected of this study (b)

Fig.2 Specimens of X. matsubarai and X. setifer
Okada and Suzuki reported a new species of genus Xiphasia, X. matsubarai, in the Sea of Japan in 1952 and identi fied its holotype (FFUM 4052) at a type locality in the Sea of Japan, off the coast of Owashi(Okada and Suzuki, 1952). The species was named for Dr. Kiyomatsu Matsubara, a famous Japanese ichthyologist. The common English name of X. matsubarai is Japanese snake blenny, a semipelagic fish, widely distributed from the Indian Ocean to the West Paci fic, in both tropical and temperate regions (Fig.1a) (Springer, 1986; Global Biodiversity Information Facility (GBIF), 2017). Several records of X. matsubarai were collected on the surface at night through light attracting fishing gear, also were found in the stomachs of tuna ( Thunnus albacares)and dolphin fish ( Coryphaena hippurus) (Springer and Smith-Vaniz, 1970; Smith-Vaniz, 1976; Smith-Vaniz, 1987). This is a poorly known nocturnal species that inhabits deep water and rises to the surface at night for predating pelagic fish (Myers,1999). Although depth record of this species occurs to 4 960 m depth from Zoological Museum, University of Copenhagen (Anonymous, 2001), according to the habits of this species may be an ocean depth of collected station. Consequently, its habitat bathymetric distribution is unclear. Genus Xiphasia is easily vulnerable to predation because of its slender body,which caused it always dives into the bottom of sand or mud when it is frightened by others (Masuda et al.,1984). These species hide in the mud during the daytime and emerge for feeding at night, which is a unique behavior for a blenniid (Myers, 1999). This species had not been reported in the South China Sea before this discovery. The specimen, with the number of GOU100852, is stored in the Center of South China Sea Fisheries Resources Monitoring and Assessment,Guangdong Ocean University.
2 MATERIAL AND METHOD
2.1 Sampling
Specimens were collected using falling net and bottom trawling. Sampling stations of this study located in the waters off of the Xisha Islands and southeastern Hainan Island (Fig.1b). Three specimens were examined. Specimen No. GOU100852 was collected by a falling net on April 23, 2016 and identi fied as X. matsubarai (Fig.2a), and its standard length was 359.54 mm. The other two specimens GOU100709 and GOU100945 (Fig.2b) were collected by bottom trawling and the standard lengths were 422.1 mm and 453.9 mm, respectively, were identi fied as X. setifer.
2.2 Morphology measurement
Specimen measurements and counting methods in this study were based on Okada and Suzuki (1952)and Smith-Vaniz (1976). Standard length and other measurements were made with an electronic Vernier calliper (Mitutoyo-IP67) with accuracy to 0.1 mm.The colour description of the specimen was described mainly from frozen specimens.
2.3 Sequence analysis
The barcode method is mainly based on COI fragments of mitochondrial DNA to identify species.Genomic DNA was extracted with a Rapid Animal Genomic DNA Isolation Kit (Sangon Biotech, Inc.,Shanghai, China) from the muscle tissue (50 mg). A polymerase chain reaction (PCR) was ampli fied by the universal primers FishF1 (5'-TCAACCAACCACAAAGACATTGGCAC-3') and FishR1 (5'-TAGACTTCTGGGTGGCCAAAGAATCA-3') (Ward et al., 2005). The PCR was performed in a 25 μL reaction mixes that included 12.5 μL of Taq PCR Master Mix(Sangon Biotech, Inc., Shanghai, China), 5 μL of DNA template, 2 μL of forward primer, 2 μL of reverse primer, and 3.5 μL of ultrapure water. PCR ampli fications were performed using a Bio-Rad T100™ Thermal Cycler (Bio-Rad Laboratories, Inc.,USA) under the following conditions: denaturation of 5 min at 95°C, followed by 33 cycles of 45 s at 94°C for denaturation, 45 s at 52°C for annealing, and 1 min at 72°C for extension, with post-extension at 72°C for 10 min and a final hold at 4°C. The PCR products were sent to Sangon Biotech (Shanghai)Co., Ltd. for purifying and sequencing. All sequences have been deposited in GenBank under accession numbers: KY643875–KY643877.
All sequences were aligned using Clustal X v1.81(Thompson et al., 1997) and then optimized manually.There were four homologous sequences of the genus Xiphasia and two out-group sequences ( Scomberomorus guttatus, Acropoma japonicum) that were download from the GenBank of NCBI. MEGA 7.0 (Kumar et al.,2016) and MrBayes 3.0b4 (Huelsenbeck and Ronquist,2001) software were used to analyze the sequences.The neighbor-joining (NJ) phylogenetic tree was constructed by the Kimura two parameter (K2P) model method with 1 000 repeated bootstraps (Felsenstein,1985; Saitou and Nei, 1987), which calculated the species and intraspeci fic genetic distance (Kimura,1980). The posterior probability values were used as support for the Bayesian topology. Log-likelihood stability was reached after approximately 1 000 000 generations; the first 500 trees were excluded as burnin values, and the remaining trees were used to compute a 50% majority rule consensus. Meanwhile, sequences were matched in the Barcode of Life Data Systems(BOLD) V3 database to verify the results.
3 RESULT
3.1 Morphological characteristics
The species X. matsubarai is distinguished by the following diagnostic characters. Body elongate eellike or ribbon-like in shape, and flanks medium flat.Head small without cirri, and bluntly rounded anteriorly. Eyes is slightly smaller and the head length is about five times longer than the length of eye diameter. There are two pairs of nostrils, the anterior nostrils very large, and the posterior nostrils very small. Mouth rather small, the opening reaches the anterior margin of the eye, the upper jaw longer in length than the lower jaw, and the upper jaw is fixed.A single row incisor-like teeth are in each jaw. On both sides of upper and lower jaws with a large canine,the canines of lower jaw longer than the canines of upper jaw which insert in the notches between canines and incisors of upper jaw. The tongue is thick, narrow and short, and the front end is free. The opercule is buried in the subcutaneous, gill opening locate above the pectoral fin base, approximately equal to the length of eye diameter. The gill opening has both a lateral and mesial flap. Smith-Vaniz and Rose (2012)suggested that these flaps probably function as a one way valve to present sand and fine sediment from entering the gill cavity. The body has no scales and lateral line has been degraded. The base of the dorsal fin is very long, dorsal fin elements XI, 96, the starting point is above the eyes. Dorsal, anal and caudal fins are connected. The origin of anal fin opposite 14th dorsal fin ray. Pectoral fins are short and rounded.Both pelvic fins are closer at the throat position.Pectoral fin elements 10, anal fin elements II, 95.Head and body grey-brown, 26 dark grey-brown bands, abdomen and lower operculum lighter yellowish grey, dorsal base long with dark grey, origin of dorsal base located above the anterior margin of pupil, black blotch on dorsal fin between the 8th and 10th dorsal spines, anal and caudal fins dark grey,pectoral and pelvic fins pale yellow.
3.2 Sequence analysis of the COI gene
The COI gene fragments of the three samples werebidirectionally sequenced, in which no insertions and deletions of the sequences were detected. It is approximately 643 bp in length, and the homologous sequences were downloaded from GenBank. The base composition of the seven COI sequences were composed of A: 24.4%, C: 25.1%, T: 33.3%, and G:17.2%. The sum of A+T (57.7%) was greater than that of G+C (42.3%). The genetic distance between the two populations was calculated using the K2P. The interspecies genetic distance of X. matsubarai was 0.0068, and the average genetic distance within the genus was 0.1406. The phylogenetic tree was constructed by neighbor-joining (NJ) and Bayesian phylogenetic analysis with two out-group sequences( S. guttatus and A. japonicum) (Fig.3). The GOU00852 sequence was clustered with JF494790 and KF489810( X. matsubarai), and the GOU00709, GOU00945,KX301896, and JQ681493 ( X. setifer) sequences were clustered, the two species were clustered into one group with high support, respectively. The sequences were submitted and compared to the data in the BOLD system; both results were consistent.
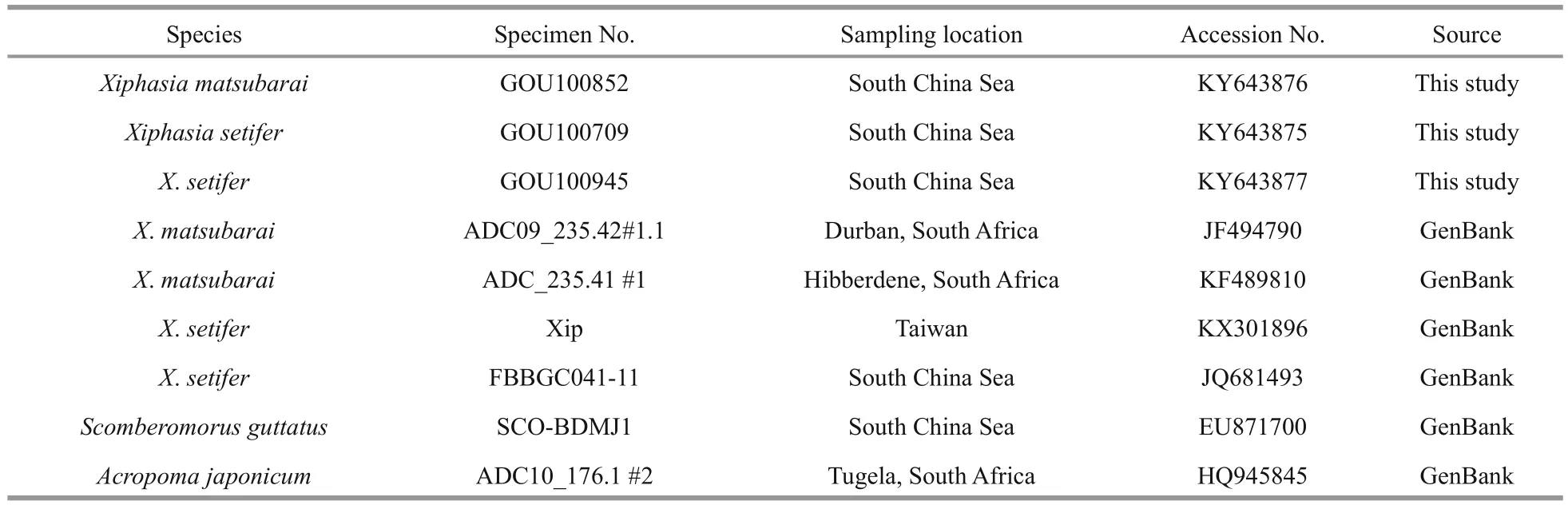
Table 1 Information of mitochondrial DNA COI sequences referenced in this study
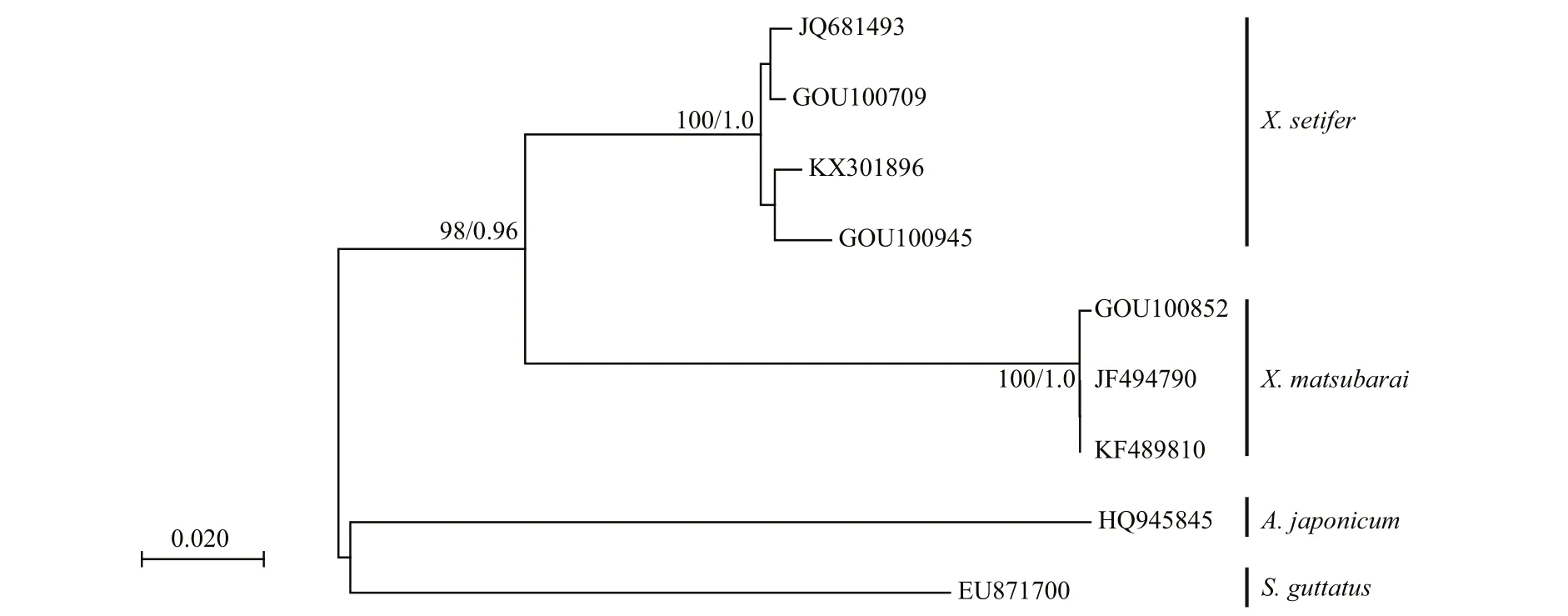
Fig.3 Phylogenetic relationships of 9 COI sequences from 4 fish species using Neighbor-joining (NJ) and Bayesian phylogenetic analyses
4 DISCUSSION
It is difficult to distinguish X. matsubarai from X.setifer as they are super ficially very similar. They both have an eel-like body, an elongated dorsal fin,and an anal fin, except there is no overlap in meristic values. To identify the species, their counts and measurements were compared to the previous records(Table 2). Smith-Vaniz and Shen reported that male adult of X. setifer have two rays extended in the middle of the caudal fin (Smith-Vaniz, 1976; Shen,1993). The caudal fin lengths of X. setifer have great variation (2.7%–21.8%), but the caudal fin lengths of X. matsubarai differ only slightly (2.1%–2.6%).There are different distinguishing features between X. matsubarai and X. setifer. The fresh specimenpigmentation of the dorsal fin is useful to distinguish these two species (Fig.4). The dorsal fins of X. matsubarai are grey-black and have a black blotch between the 8th and 10th spines, whereas X. setifer have dorsal fins that are brown-green with a black spot in the middle of the 7th spine, a large black blotch
between the 12th and 16th spines, and a faint trace of yellowish bands in front of it. Comparing these two species, the dorsal fin origin of X. matsubarai is positioned more posteriorly (above the anterior margin of the pupil), while in X. setifer it begins slightly in front of the anterior edge of the bony orbit.The anal fin origin of X. matsubarai relative to the position of the dorsal fin is the 14th ray, but in X. setifer it is the 18th ray. The number of pectoral fin elements of X. matsubarai is 10 or 11, but the number of elements of X. setifer is 12 to 14.
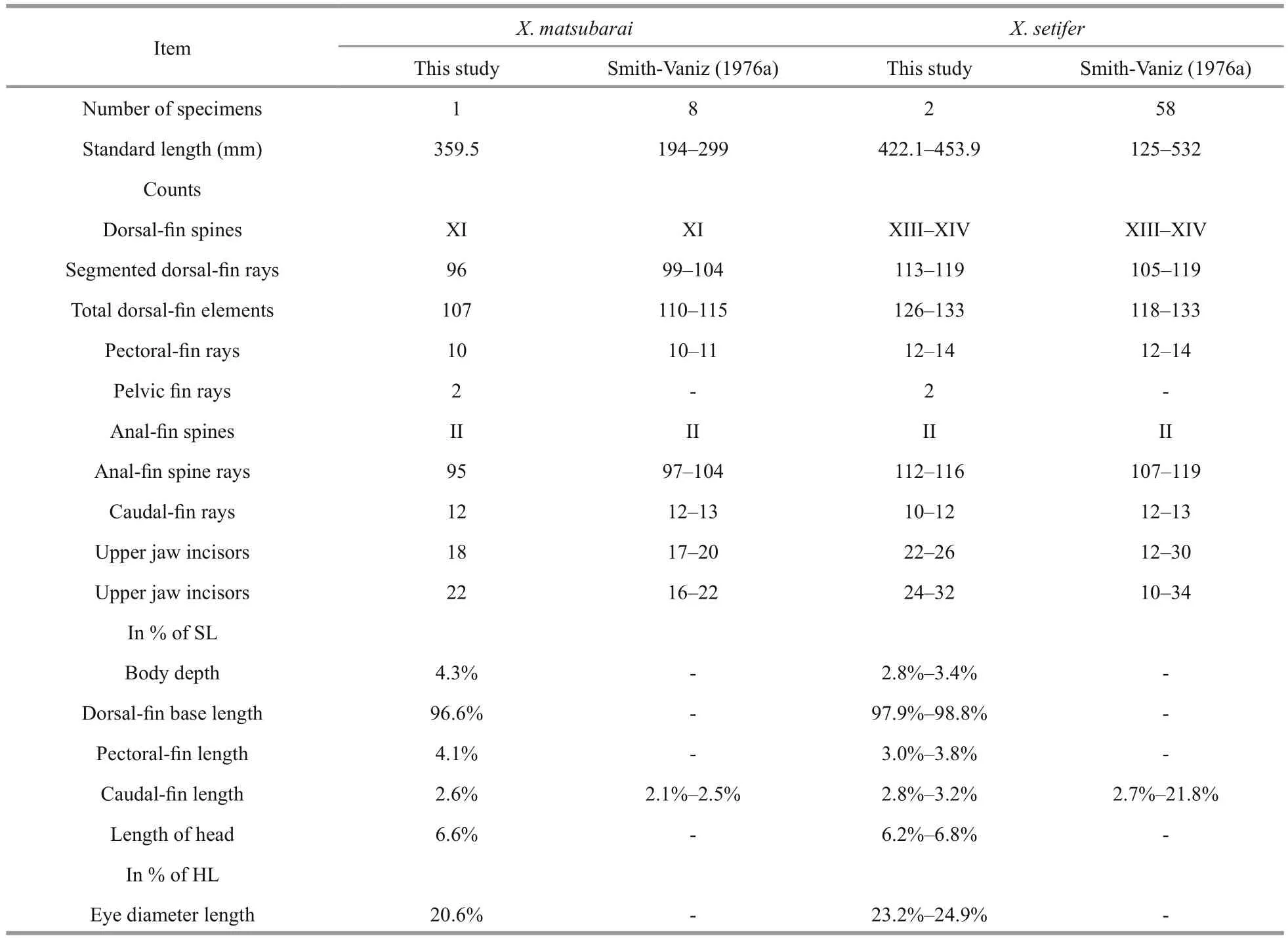
Table 2 Comparison of counts and measurements of X. matsubarai and X. setifer
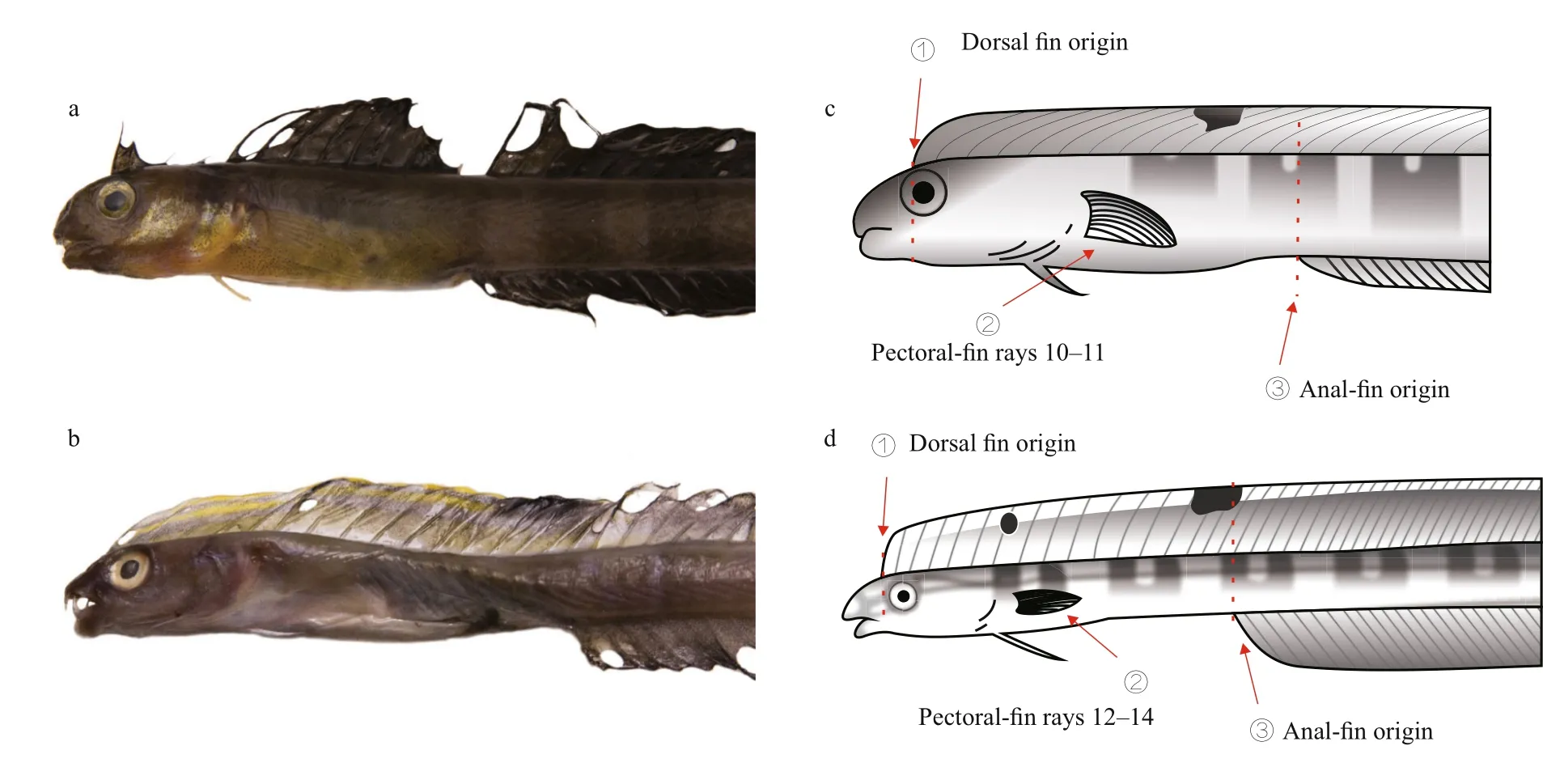
Fig.4 Comparison of the front body of X. matsubarai (a, c) and X. setifer (b, d)
Using DNA barcoding of the COI sequence, fish species can be identi fied effectively. In this study,firstly we con firmed the morphology of X. matsubarai and then combined the results with those of the DNA barcoding molecular methods, which con firmed the species identity. In comparison with the homologous COI sequences from GenBank, the X. matsubarai sequences of this study were clustered with two sequences of X. matsubarai, and the identity showed a 100% match. The results are the same in the BOLD system. The K2P model was used to analyse the average genetic distance of the species in X. matsubarai, which was 0.006 8, and the average genetic distance of genus Xiphasia was 0.140 6. The genus genetic distance was approximately 20.68 times higher than that of the species, which was signi ficantly higher than values found in Hebert’s thesis, where species and interspeci fic distances were 10-times greater (Hebert et al., 2003). The above results have further identi fied a new record species of X. matsubarai.
5 CONCLUSION
By comparing the results, the morphology and counts of the specimens’ characteristics, they are similar to those described by Okada, Smith-Vaniz and Nakabo (Okada and Suzuki, 1952; Smith-Vaniz,1976; Nakabo, 2013). However, both the count of total dorsal fin elements and the total anal fin elements of X. matsubarai in the South China Sea of this study are less than those in the South Africa Sea. The result of DNA barcoding analysis support this species as X. matsubarai, which has not been previously reported from the South China Sea. A key to the species of the genus Xiphasia is also given.
Key to the species of the genus Xiphasia
1a. Pectoral fin 10–11; dorsal fin spines XI; dorsal fin rays 96–104; anal fin spines II, anal fin rays 95–104; origin of dorsal fin located above anterior margin of pupil, the origin of anal fin located at 14th dorsal fin ray……….............................................… Xiphasia matsubarai Okada & Suzuki, 1952
1b. Pectoral fin 12–14; dorsal fin spines XIII–XIV;dorsal fin rays 105–119; anal fin spines II, anal fin rays 107–119; origin of dorsal fin located above anterior margin bony orbit; origin of anal fin located at 18th dorsal fin ray.…......................……… Xiphasia setifer Swainson, 1839
6 DATA AVAILABILITY STATEMENT
The COI sequence data that support the findings of this study have been deposited in GenBank with the accession codes KY643875–KY643877. The species of Xiphasia matsubarai distribution data that support the findings of this study are available from Global Biodiversity Information Facility (GBIF), http://www.gbif.org/species/2395301/.
7 ACKNOWLEDGEMENT
The authors gratefully thank Dr. LIN H. D., for the suggestions of experiment and data analysis. We also thank Prof. KANG B. and Dr. HOU G. for offering assistance with writing the paper. Thanks are also to anonymous referees and the editors for their valuable comments, which have greatly improved this paper.
杂志排行
Journal of Oceanology and Limnology的其它文章
- Editorial Statement
- Recent insights into physiological responses to nutrients by the cylindrospermopsin producing cyanobacterium,Cylindrospermopsis raciborskii*
- Response of Microcystis aeruginosa FACHB-905 to different nutrient ratios and changes in phosphorus chemistry*
- In fluence of light availability on the speci fic density, size and sinking loss of Anabaena flos- aquae and Scenedesmus obliquus*
- Application of first order rate kinetics to explain changes in bloom toxicity—the importance of understanding cell toxin quotas*
- Regime shift in Lake Dianchi (China) during the last 50 years*
